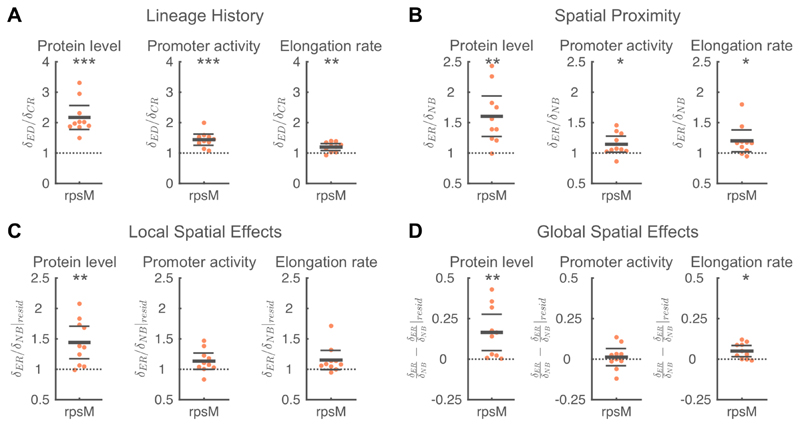
Figure 6. Analyses of factors that contribute to spatial correlations in metabolism.
A) Shared lineage history leads to similarity in RpsM protein levels (left), rpsM promoter activity (middle), and cell elongation rate (right). In all cases, the phenotypic difference between a focal cell and an equidistant cell (δED) is significantly larger than the phenotypic difference between a focal cell and its closest relative (δCR). B) Spatial proximity leads to similarity in RpsM protein levels, rpsM promoter activity, and cell elongation rate. In all cases, the phenotypic difference between a focal cell and an equally-related cell (δER) significantly is larger than the phenotypic difference between the focal cell and one of its neighbors (δNB). C) The similarity in RpsM protein levels is partly due to local spatial effects. For RpsM protein levels, the difference in residuals between a focal cell and an equally-related cell (δER|resid) is significantly larger than the difference in residuals between the focal cell and one of its neighbors (δNB|resid). D) Global spatial effects lead to similarity in RpsM protein levels and cell elongation rate. A-D) Each point corresponds to a microcolony with 117-138 (mean=128) cells, points are horizontally offset. Thick horizontal lines indicate mean, thin lines 95% confidence intervals. Dashed lines indicate the expected value under the null hypothesis. Null hypothesis rejected with: *p<0.05, **p<0.01, ***p<0.001, t-test, n=10. See Figure S10 for a chromosomal rpsM reporter in Salmonella Tm. See also Figure S5-7.