Figure 5.
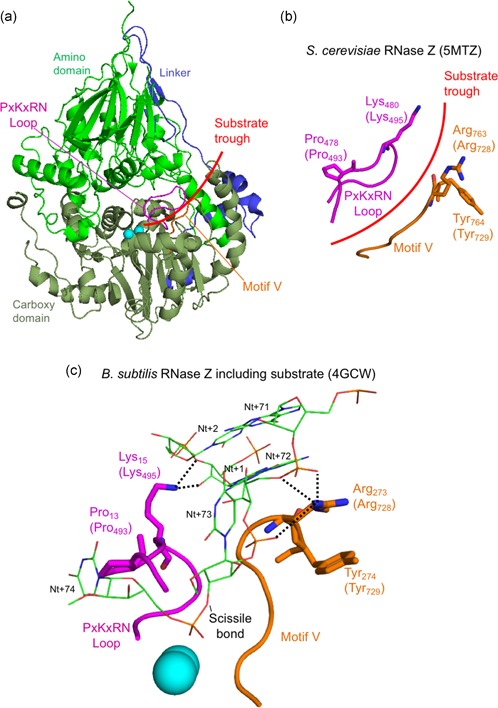
In silico analysis of disease‐related ELAC2 substitutions p.Pro493Leu and p.Tyr729Cys. (a) Structure overview of S. cerevisiae Trz1. Amino and carboxy domains and linker are colored as in previous figures. The curved arrow indicates the presumed substrate trough. Metal ions which mark the active site are shown as blue spheres. PxKxRN and Motif V loops are shown in magenta and orange, respectively. (b) Detailed view of PxKxRN loop (magenta) and Motif V loop (orange) in S. cerevisiae Trz1. The conserved basic residues Lys in the PxKxRN loop and Arg in the Motif V loop are shown as sticks, with H. sapiens equivalent residues given in brackets. Red arc illustrates the path of substrate acceptor stem and 3′ trailer through the presumed substrate trough. (c) Detailed view of PxKxRN loop (magenta) and Motif V loop (orange) in B. subtillis with pre‐tRNA substrate (Note: The B. subtillis structure displays similar folds and relative orientations, validating its use for modeling ELAC2 (RNase ZL) structure including the carboxy domain and the active site. The B. subtilis RNase Z structure 4GCW (Pellegrini et al., 2012) is the only available structure of an enzyme—pre‐tRNA substrate complex). The pre‐tRNA substrate acceptor stem is clamped by polar contacts with K in the PxKxRN loop (Lys495 in H. sapiens, Lys480 in S. cerevisiae, Lys15 in B. subtilis) and Arg in the Motif V loop (Arg728 in H. sapiens, Arg763 in S. cerevisiae, Arg273 in B. subtilis), illustrated with bold dashed lines. A polar substrate acceptor stem clamp consists of Lys15 contacts with 2′ and 3′ O's of ribose +1 and Arg273 contacts with two backbone phosphate O's on nt 72 and one on nt 73. Counterintuitively, the basic R‐groups that form the substrate clamp do not point toward each other; both are oriented toward the right, but two polar contacts extend to the right from Lys15 while three polar contacts by Arg273 extend to the left toward substrate
