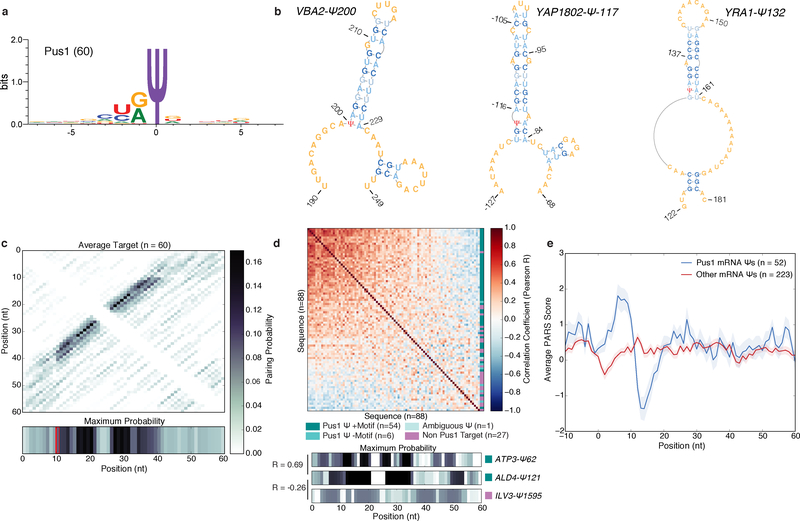
Figure 3: A Structural Motif Associated with Pus1 mRNA Targets in Yeast.
a) The sequence motif surrounding n=60 high confidence PUS1-dependent mRNA Ψs, generated with WebLogo 3.5. b) MFE structures for VBA2-Ψ200 (left), YAP1802-Ψ-117 (middle) YRA1-Ψ132 (right). c) A heatmap of the average pairing probability matrix from RNAfold (upper), and the average maximum pairing probability for each base (lower) for n=60 high-confidence Pus1 targets. d) A heatmap of pairwise correlation coefficients (Pearson R) between the arrays of maximum pairing probabilities for each mRNA site (n=88 Ψs with genetic evidence for Pus1-dependence in vivo). In vitro modified mRNA Ψs with a stem-loop motif (dark teal, n=54 seqeunces), in vitro modified without a stem-loop motif (medium teal, n=6 sequences), with ambiguous in vitro data (light teal, n=1 sequence), and not modified in vitro (purple, n=27 sequences). Rows and columns are ordered by the sum of R values across the row/column. Indicated on the right is the classification of each sequence (upper). The maximum pairing probability and correlation values for 3 sequences (lower). e) Average PARS score ±SEM for our high confidence Pus1 mRNA substrates (blue, n=52 targets), and all other mRNA Ψs (red, n=223 targets) identified in log phase and high density for which there is PARS data.