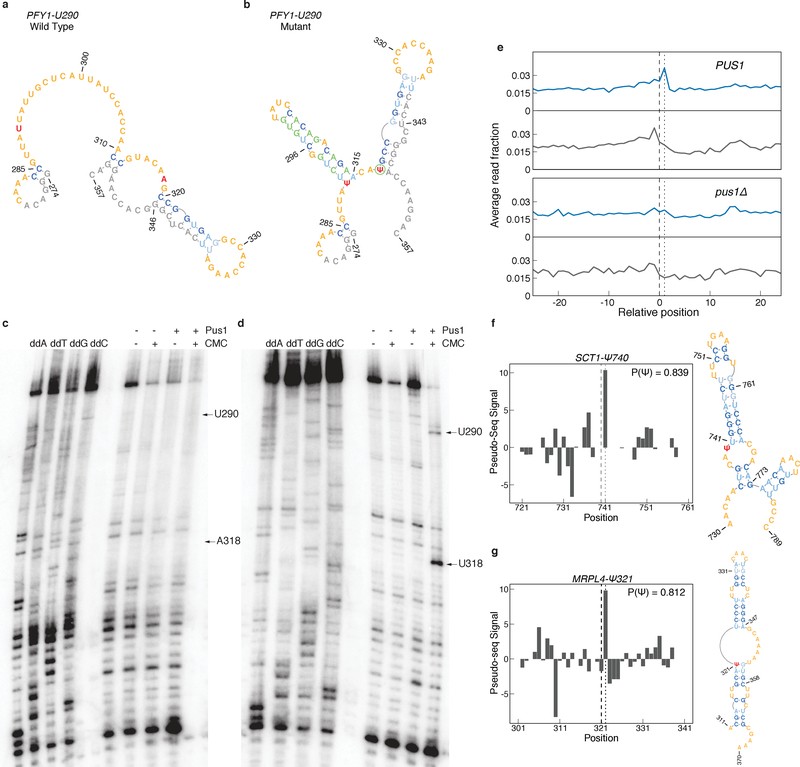
Figure 6: The Pus1 Structural Motif is Sufficient for Pseudouridylation.
a-b) MFE structures for a region surrounding (a) wild type, or (b) mutant PFY1-U290. Target Ψ nucleotides (red), adapter, padding, and T7 sequences (gray), and mutated nucleotides (green) are indicated. c-d) Primer extension gels of (c) PFY1-U290 wild type, and (d) PFY1-U290 mutant sequences. Positions of U290 and A/U318 are indicated. Gels are representative of n=4 replicates. Uncropped gel images can be found in Supplementary Figure 7a,b. e) Metaplot of RT stops for sites predicted to be Pus1 targets with P(Ψ) > 0.8, in +CMC libraries (blue) and −CMC libraries (grey) from high OD in vivo Pseudo-seq data. PUS1 panels show the aggregated reads from knockout libraries for pus2Δ,3Δ,4Δ,5Δ,6Δ,7Δ, and 9Δ. f-g) Pseudo-seq signal from the pooled PUS1 reads, and predicted secondary structure for a putative Pus1 target at (f) position SCT1-Ψ740 and (g) MRPL4-Ψ321.