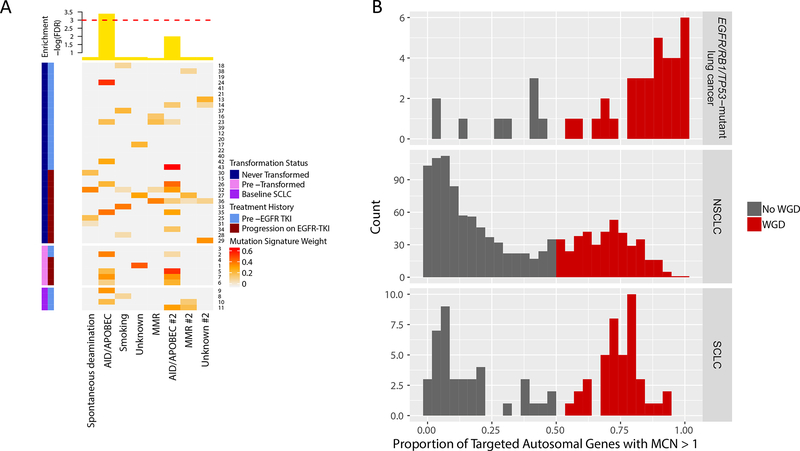
Figure 4.
AID/APOBEC mutation signature and whole genome doubling (WGD) in EGFR/RB1/TP53-mutant lung cancers. (A) Mutation signature analysis identified significant enrichment of AID/APOBEC in pre-transformed SCLC compared to never transformed. Heatmap is calculated based on weights [0,1] measuring how strongly a mutation signature is represented in a given sample. Top annotation plots significance as -log(FDR) for enrichment of a mutation signature in pre-transformed SCLC. The dashed red line corresponds to an FDR of 0.05. (B) The frequency of WGD in lung cancer with concurrent EGFR/RB1/TP53 mutations is higher compared to all NSCLC and SCLC. Plots show the distribution of the proportion of autosomal genes with major copy number of at least 2 in lung cancers with concurrent EGFR/RB1/TP53 mutations, NSCLC, and SCLC respectively. Using a cutoff of 50%, the proportion of samples with WGD is colored in red, corresponding to a WGD frequency of 80% in lung cancer with concurrent EGFR/RB1/TP53 vs 34% in NSCLC (p < 5×10−9) vs 51% in SCLC (p < 0.002).