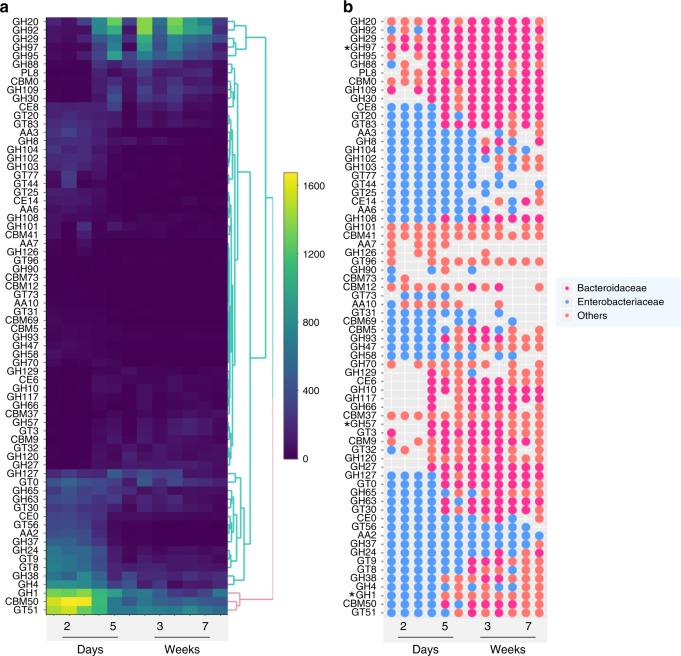
Fig. 10.
The abundance, distribution and predicted bacterial taxonomy of fecal CAZy families. a Heatmap depicting the normalized distribution of CAZy enzymes over time, including only differential abundant enzymes (n = 70). b The most abundant bacterial family predicted to produce the corresponding CAZy enzymes. Dots indicate that the predicted family was either Enterobacteriaceae (blue), Bacteroidaceae (pink), or Others (orange). The absence of a dot indicates that either our inability to predict the bacterial origin or the CAZy family was not detected in a particular calf at a particular time point. Enzymes families GH1 which includes lactase (EC 3.2.1.108), GH97 which includes glucoamylase (EC 3.2.1.3) and α-glucosidase (EC 3.2.1.20)), and family GH57, which includes α-amylase (EC 3.2.1.1) are highlighted with asterisks. Abbreviations: GH, glycoside hydrolases; GT, glycosyltransferases; PL, polysaccharide lyases; CE, carbohydrate esterases; AA, auxiliary activities; CBM, carbohydrate-binding modules. Families CBM0, CE0, and GT0 refer to unclassified CBMs, CEs and GTs, respectively, in CAZy database download 07152016. Differential abundance analysis with DESeq2 revealed that 71 CAZy families significantly changed over time in dairy calves, however, we failed to retrieve the bacterial origin of one CAZy family, GT80, and thus there are a total of 70 CAZy families shown here. Source data are provided as a Source Data file