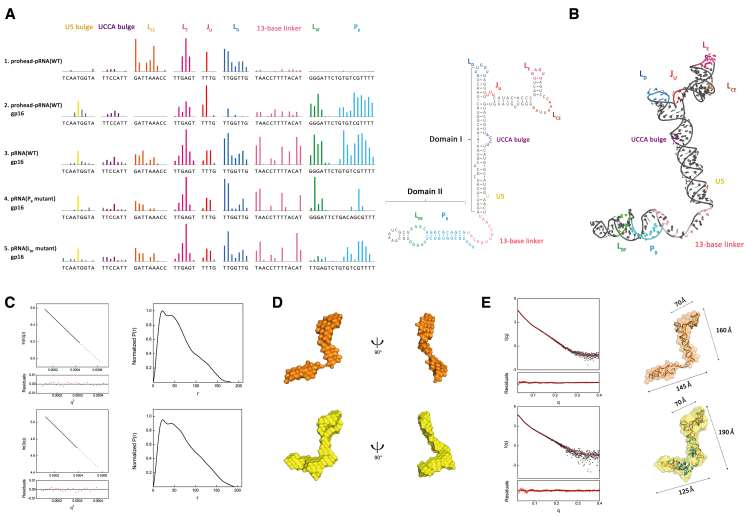
Figure 2.
The ATPase gp16 binds to the pRNA and induces conformational change. (A) SHAPE analysis shows that gp16 specifically recognizes pRNA domain II. Upward colored bars represent reduced SHAPE activity upon prohead-bound pRNA (row 1), gp16 binding to the prohead-pRNA (row 2) and free wild-type pRNA (row 3), a PB mutant where the sense (UGUCGU) and anti-sense (ACAGCG) strand are switched (row 4) and a LBF mutant (GGGAT to TTGAG) (row 5). Residues are indicated on the X-axis. Coloring of pRNA secondary structure is consistent with SHAPE signal. (B) Schematic representation of all protected residues on the reconstructed 3D pRNA model. The model contains the previously solved pRNA domain I structure (PDB code: 3R4F, 4KZ2), a helical region grafted from the hexamer pRNA model (PDB code: 1L4O) and a 13-base linker constructed in COOT. Coloring is consistent between the 3D model and SHAPE signal. Protected sites on LCE, LE, JU and LD of the RNA ring are labeled in orange, magenta, red and navy, respectively, showing intra-RNA interactions. The UCCA bulge and U5 on stem A are shown in purple and yellow, respectively. The 13-base linker is shown in light pink. The signal from LBF, PB on pRNA domain II are shown in green and cyan. (C) Guinier plot (left) and normalized P(r) analysis (right) of the pRNA (upper series) pRNA-gp16 RNP (lower series). (D) Low-resolution bead models calculated by DAMMIF from SAXS data. The pRNA (upper series) is shown in orange, and RNP is shown in yellow. (E) Atomic models of pRNA docked inside the SAXS bead models. The pRNA atomic structure theoretical solution scattering curve (red) is compared to the experimental scattering curves (black) by FoXS. Coloring of pRNA (upper series) and RNP (lower series) bead model is consistent with that in panel (D). Dimensions of RNA ring, stem A and domain II are 70, 160 and 145 Å in pRNA. The three motifs of RNP have dimensions of 70, 190 and 125 Å. The angle between pRNA domain II and A stem is 92°.