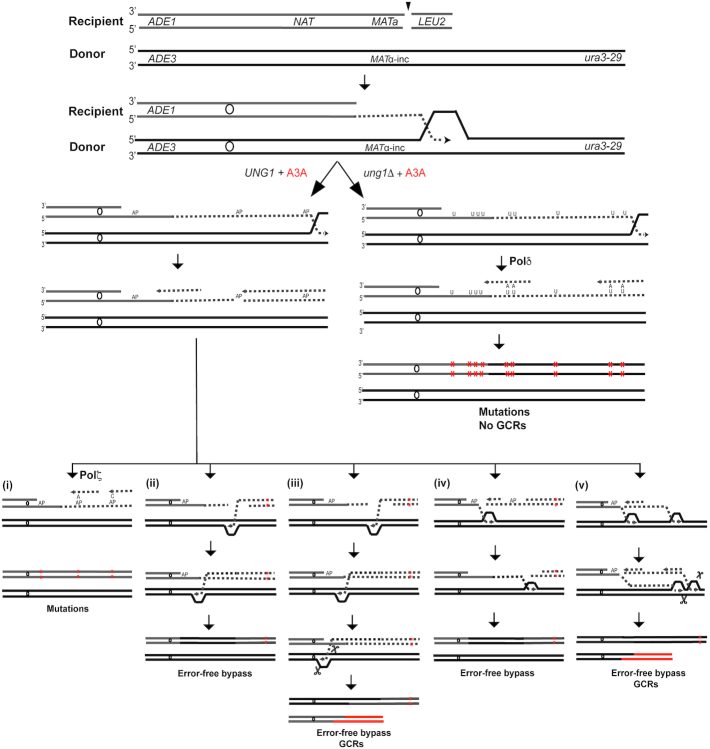
Figure 7.
Formation of A3A–induced mutation clusters and GCRs during BIR. DSBs are initiated at MATa. 5′-3′ resection of DSB ends occurs followed by invasion of 3’ ssDNA ends into the homologous chromosome and initiation of DNA synthesis. A3A deaminates cytidines in ssDNA accumulated during leading strand BIR synthesis, which leads to the formation of uracils (U). Mutations (red asterisks) result from incorporation of adenines (A) across from uracils during lagging strand BIR synthesis in ung1Δ (right). In UNG1 cells, uracil is excised by uracil glycosylase leading to the formation of abasic (AP) site (left). During lagging strand synthesis, Pol δ stalls at AP site, which can lead to the following outcomes: (i) Pol ζ -mediated error-prone bypass of AP site giving rise to C to T or C to G mutations; (ii) Breakage of AP site followed by invasion of 3’ stalled ssDNA end into homologous chromosome followed by initiation of DNA synthesis leading to the error-free bypass of AP site; (iii) DNA synthesis using homologous chromosome is initiated similar to (ii), but interrupts due to collision at some replication obstacle (e.g. centromere) resulting in half-crossover and GCRs; (iv) Breakage at AP site leads to the invasion of another (centromere-proximal) 3’-ssDNA end into the homolog followed by DNA synthesis proceeding in the direction of telomere and resulting in error-free bypass of AP sites or (v) Collision of error-free bypass synthesis with BIR bubble leading to interruption of DNA synthesis and formation of GCRs.