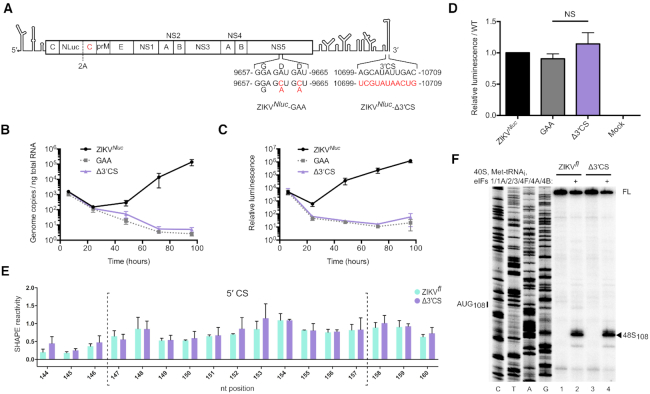
Figure 2.
Genome circularization is required for ZIKV replication. (A) Schematic illustration of ZIKVNluc. The first copy of the capsid gene is fused to a Nluc reporter and is separated from a second capsid sequence and the rest of the polyprotein by the FMDV 2A StopGo (40). ZIKVNluc-GAA and ZIKVNluc-Δ3′ CS polymerase mutations are indicated. (B andC) Time course of (B) ZIKV genome copy number quantified by qPCR and (C) luciferase activity normalized to total protein amount after electroporation of capped ZIKVNluc RNA into Vero cells. Data are mean ± SEM for three independent experiments. (D) Luciferase activity normalized to total protein amount at 6 h post-electroporation relative to the WT control (full time course shown in C). Data are mean ± SEM of three independent experiments. ZIKVNluc-GAA and ZIKVNluc-Δ3′ CS mutants were compared by Mann–Whitney test (P = 0.40). NS, not significant. (E) SHAPE reactivity in the 5′ CS of ZIKVfl (green) and ZIKVfl-Δ3′ CS (purple). Nucleotides within the 5′ CS are enclosed by dotted brackets. Data are normalized SHAPE reactivities from three experiments, mean ± SD at each base. SHAPE reactivities in the 5′ CS region of ZIKVfl and ZIKVfl-Δ3′ CS mutant were compared by the Student's t-test and determined not to be significant. P-values are shown in Supplementary Table S2. (F) Toeprinting analysis of 48S complex assembly on capped ZIKVfl and ZIKVfl-Δ3′ CS RNA. Selected codons are labelled on the left and toeprints caused by 48S complex assembly are marked with a closed arrowhead on the right. FL, full-length.