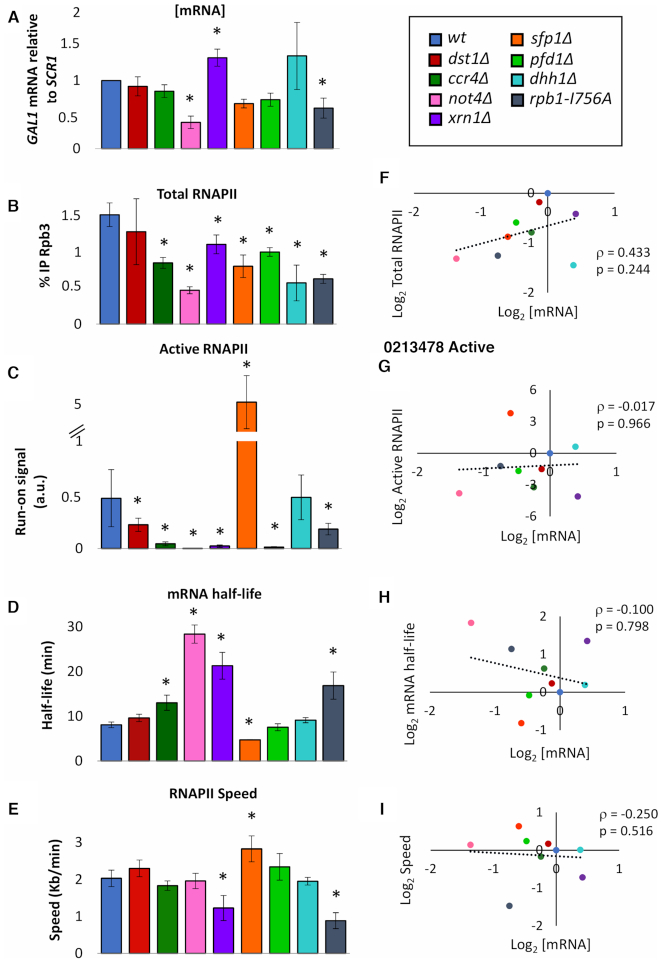
Figure 1.
GAL1 mRNA levels resists most perturbations of synthesis and decay. (A) GAL1 mRNA levels in exponential cultures of the indicated mutant strains. Apparent [mRNA] was calculated relative to SCR1 by RT-qPCR. (B) Total RNAPII occupancy as measured by anti-Rpb3 ChIP. Each bar represents the average value of four amplicons across the GAL1 gene. (C) Active RNAPII as measured by the incorporation of radioactive 32UTP (transcriptional run-on). The bars represent the average value of four probes across the GAL1 gene. (D) GAL1 mRNA as measured by RT-qPCR before and after shutting off transcription by glucose. Half-lives were determined by calculating the time it takes for half of the initial mRNA to be degraded. (E) RNAPII speed as measured by anti-Rpb3 ChIP in the long GAL1p-YLR454w gene before shutting off transcription, and three times afterwards. Values were normalized to time 0, and RNAPII speed was calculated. All bars represent mean and standard deviation of three biological replicates (* if p < 0.05 in a Student’s t-test). (F–I) To the right of each of the previous graphs, we have represented the corresponding variable with respect to apparent [mRNA]. We never found any significant correlation using a Spearman’s test (ρ represents the Spearman coefficient and the p, the p-value).