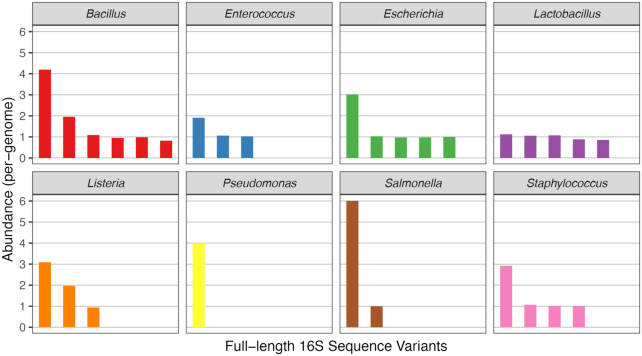
Figure 2.
Abundances of full-length 16S rRNA gene amplicon sequence variants (ASVs) detected in the Zymo mock community, scaled by the genomic abundance. Twenty-nine distinct ASVs were detected by our long-read amplicon sequencing methodology in the Zymo mock community. Each ASV was grouped into a bin corresponding to eight bacterial strains in the mock community on the basis of its taxonomic assignment. The abundance of each ASV was divided by the genomic abundance of the mock community strain from which it originated (see Materials and Methods), and the normalized abundance of each ASV is plotted on the y-axis. Panels correspond to the 8 bacterial strains in the Zymo mock community, most of which were associated with multiple unique ASVs, i.e. distinct alleles from different copies of the rrn operon. No other ASVs were detected.