Figure 5.
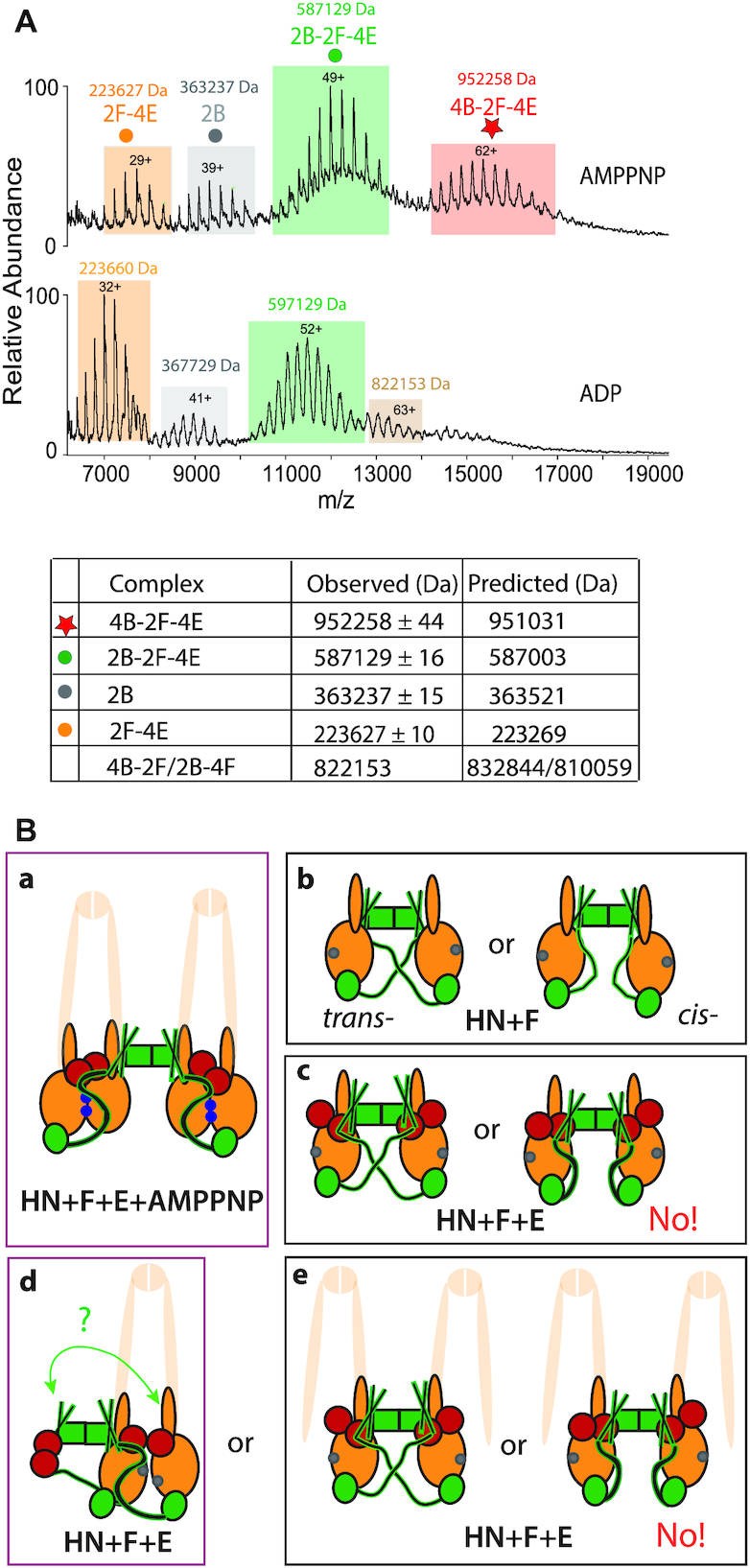
Dimers of dimers are formed with full length MukB dimers, MukEF and AMPPNP. (A) Native mass spectra of complexes formed with MukBEF in the presence of AMPPNP (top), or ADP (bottom). The predicted and observed masses are tabulated below the graphs. The proteins were at concentrations 5 μM B, 2.5 μM F and 5 μM E. The small population of complexes (mass 822153 Da, beige) observed in the ADP sample may reflect a presence of 4B-2F or 4B-2F-E2 complexes (predicted mass 832843.68 Da and 891578 Da, respectively). (B) Schematics of the architectures demonstrated or inferred from the biochemical analyses. The extrapolation from HN to full length MukB dimers is cartooned by showing the remainder of MukB as semi-transparent. Panel (a), MukBEF-AMPPNP- and head engagement-dependent dimer of dimers. (b) Alternative possible architectures of HN + MukF. (c) As (b) in the presence of MukE. The data provide evidence for the trans-configuration shown on the left. Panels d and e, show possible configuration of 2HN-2F-4E dimers in the presence of ADP/absence of head engagement. Note that the architectures in d and e-left are topologically identical if both necks engage with a 4-helix bundle (green arrow; see Figure 4D), although if part of a full-length MukB dimer as indicated, would be a dimeric MukBEF complex (d) or dimer of dimer complex (e). ATP-bound ATPase active sites denoted as blue dots on the heads and ADP-bound or nucleotide-unbound as grey dots.
