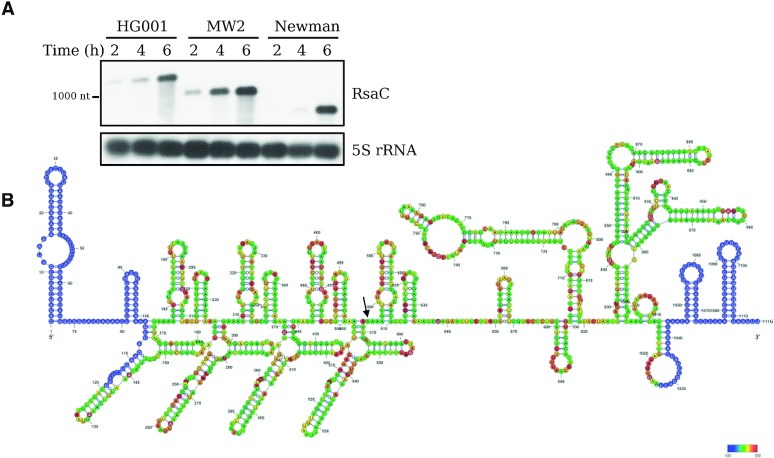
Figure 1.
RsaC is a large and highly structured sRNA. (A) Northern blot analysis of RsaC sRNA in three different S. aureus strains (HG001, MW2 and Newman). Total RNA was extracted after 2, 4 and 6 h of growth in BHI at 37°C. 5S rRNA was used as loading control. Data are representative of three independent experiments. (B) Secondary structure model of RsaC. The structure of RsaC (from HG001 strain) was mapped using selective 2′-hydroxyl acylation by benzol cyanide and the reactivity was analyzed by primer extension (SHAPE). Data were analyzed, and each modification was quantified using QuSHAPE software. The secondary structure was drawn using RNA structure and VARNA considering the reactivity of each ribose. The regions, which were not mapped, are shown in blue. The color code for each reactivity is given on the right side of the secondary structure model, red is for the highest reactivity while green is for the lowest reactivity. A black arrow indicates the +1 of RsaC544.