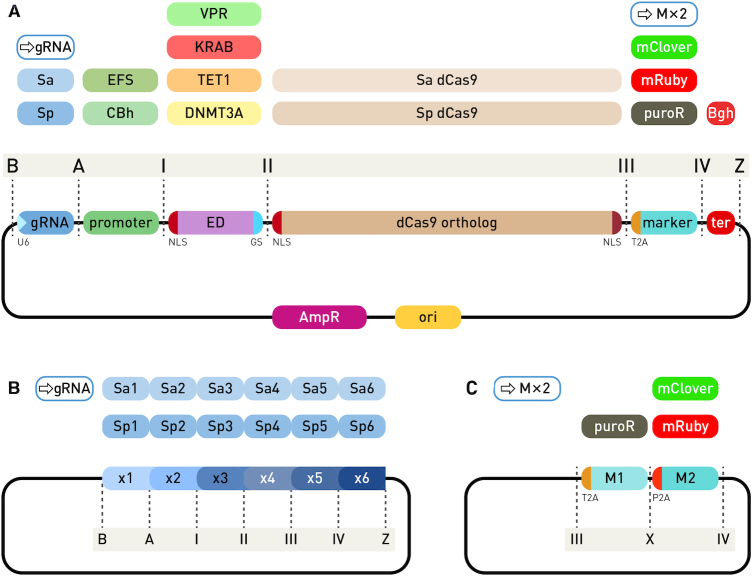
Figure 3.
Modular system for CRISPR/dCas9-based epigenetic editing and direct gene regulation. (A) Individual modules of an expression cassette for N-terminal dCas9 fusions are assembled into the backbone vector using Golden Gate cloning with the BsaI type IIS restriction enzyme. The backbone plasmid has ends compatible with ‘B’ and ‘Z’ type module ends. The first position (‘B’ to ‘A’) receives a gRNA expression module with SaCas9 or SpCas9 scaffold, either containing a pre-inserted variable gRNA region, an empty module for gRNA cloning with red-white selection or a multi-guide module for a second step insertion of up to six gRNAs. Next position (‘A’ to ‘I’) is for insertion of a eukaryotic promoter, followed by the effector domain (‘I’ to ‘II’) containing an N-terminal NLS and a short G4S linker to a dCas9 ortholog (‘II’ to ‘III’), followed by a selection marker (fluorescence or antibiotic resistance, ‘III’ to ‘IV’) linked via the self-cleaving T2A peptide, which can be substituted with the dual-marker system. Finally, a module for eukaryotic transcription terminator is inserted between ends ‘IV’ and ‘Z’. (B) The multi-guide system accepts up to six gRNA modules for either dSpCas9 or dSaCas9 protein. Individual modules require the variable part of gRNA pre-cloned for the second step of assembly, facilitated by the type IIS restriction enzyme Esp3I and red–white selection. (C) The dual marker system enables addition of both antibiotic resistance and a fluorescent protein. The dual modules have T2A and P2A self-cleaving peptides for equimolar expression with dCas9.