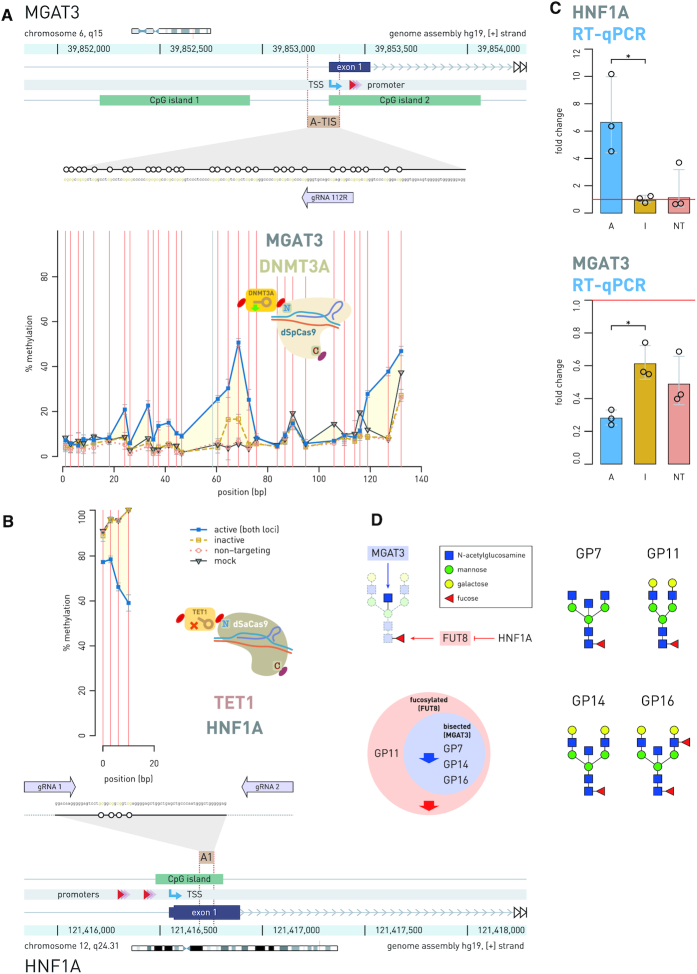
Figure 6.
Simultaneous epigenetic manipulation of the MGAT3 and HNF1A loci using DNMT3A-dSpCas9 and TET1-dSaCas9 showed an effect on total N-glycome in BG1 cells. (A) Thirty CpG sites within 200 bp wide putative regulatory region, residing in CpG island 2 of the MGAT3 promoter and centered about 100 bp upstream from the TSS, were targeted with DNMT3A-dSpCas9 using a single gRNA. (B) At the same time, four regulatory CpG sites in the first exon of the HNF1A promoter was targeted with TET1-dSaCas9 using two gRNAs. (C) Induced hyper- and hypo-methylation resulted in significant change in gene transcript level: MGAT3 showed 0.28-fold change (P = 0.004) and HNF1A showed 6.64-fold change (P = 0.004). A, dCas9 fusions with active catalytic domain; I, dCas9 fusions with inactive catalytic domain; NT, active dCas9 fusions with non-targeting gRNA. (D) The MGAT3 gene downregulation led to a statistically significant decrease (P = 0.005 compared to inactive DNMT3A-dSpCas9; P = 0.032 compared to mock-transfected cells) of the glycan structures with bisecting GlcNAc (GP7, GP14 and GP16). The HNF1A gene upregulation resulted in decrease (P = 0.005 compared to inactive TET1-dSaCas9, P = 0.028 to mock-transfected cells) of core fucose in the glycan structures with and without bisecting N-GlcNAc, probably through a known effect of HNF1A on the fucosyltransferase FUT8. Effects of HNF1A on fucosylation were represented by the sum of abundance of all complex core-fucosylated structures that are products of FUT8: GP7, GP11, GP14, GP16. Analogously, the effect of GNT-III enzyme, encoded by MGAT3, was represented by the sum of abundance of all structures containing bisecting N-GlcNAc: GP7, GP14, GP16. Thick blue and red arrows on the Venn diagram show the direction of the glycan change (a decrease of certain glycan structures in the total N-glycome of BG1 cells). * P < 0.05.