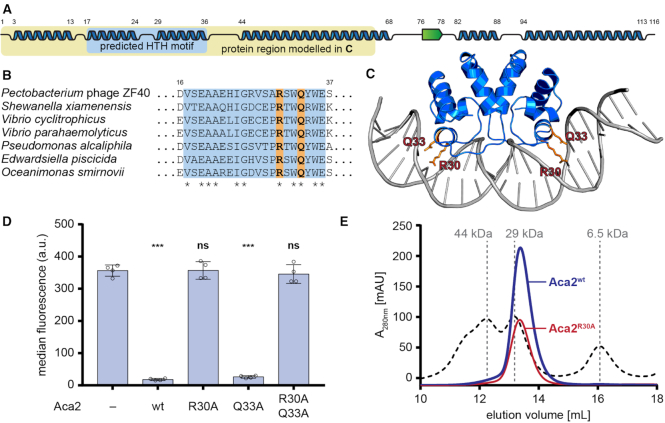
Figure 4.
Aca2 is a dimeric HTH protein. (A) Predicted secondary structures of Aca2 from phage ZF40, with α-helices displayed as blue ribbons and a β-strand as a green arrow and their amino acid positions indicated by numbers. The predicted HTH motif is highlighted in light blue. The yellow box highlights the part of the protein used for modelling in panel (C). (B) Alignment of Aca2 HTH motifs from various bacterial species, with highly conserved residues indicated by an asterisk. Residues mutated in Aca2 point mutants are highlighted in orange. Numbers indicate amino acid positions in the ZF40 homolog. (C) An Aca2 dimer (region highlighted in panel A) modeled with a DNA template, based on the published structure of MqsA in complex with DNA. The residues R30 and Q33 are highlighted. (D) Median eYFP fluorescence, as measured during a reporter assay, for different Aca2 point mutants. Data presented are the mean ± standard deviation of four biological replicates and statistical significance was calculated by one-way ANOVA with Dunnett's Multiple Comparisons Test (***P < 0.001, ns: P > 0.05). (E) SEC elution profile of wild-type Aca2 (blue) and Aca2R30A (red), with size standards indicated by dashed lines for comparison.