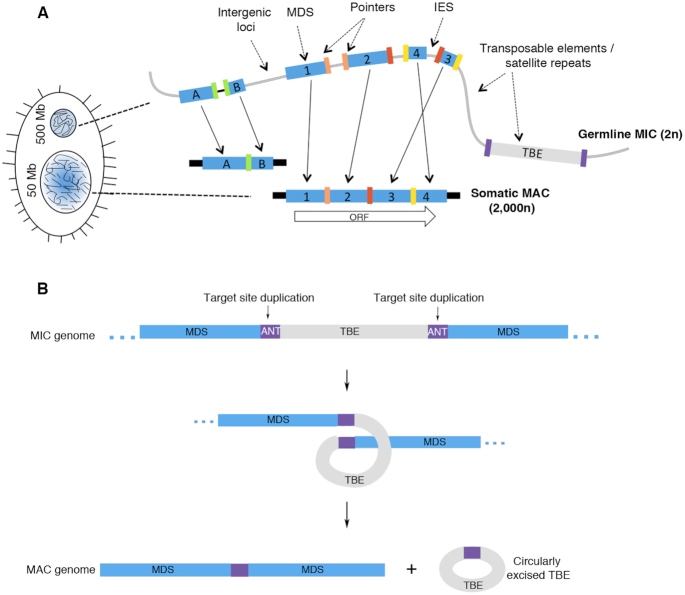
Figure 1.
Genome rearrangements in Oxytricha. (A) Cartoon illustrating Oxytricha’s dual genome architectures. The germline micronuclear genome comprises of long chromosomes that mostly contain germline-limited sequences (gray line). Somatic macronuclear nanochromosomes contain only MDSs (blue rectangles). MDSs belonging to two different MAC nanochromosomes are labeled with letters or numbers. Short colored blocks with matching color denote pointers, short direct repeats that are present downstream of the nth MDS and upstream of (n + 1)th MDS. Black blocks at the ends of nanochromosomes denote telomeres, added de novo during genome rearrangement. The chromosome copy numbers are represented by 2n and ∼2000n for the MIC and MAC genomes, respectively. (B) A model for circular TBE elimination during rearrangement. Elimination of transposon-like sequences called TBEs (in gray) via precise excisions at the ANT target site duplication (purple) and subsequent circularization based on previous work is shown (17).