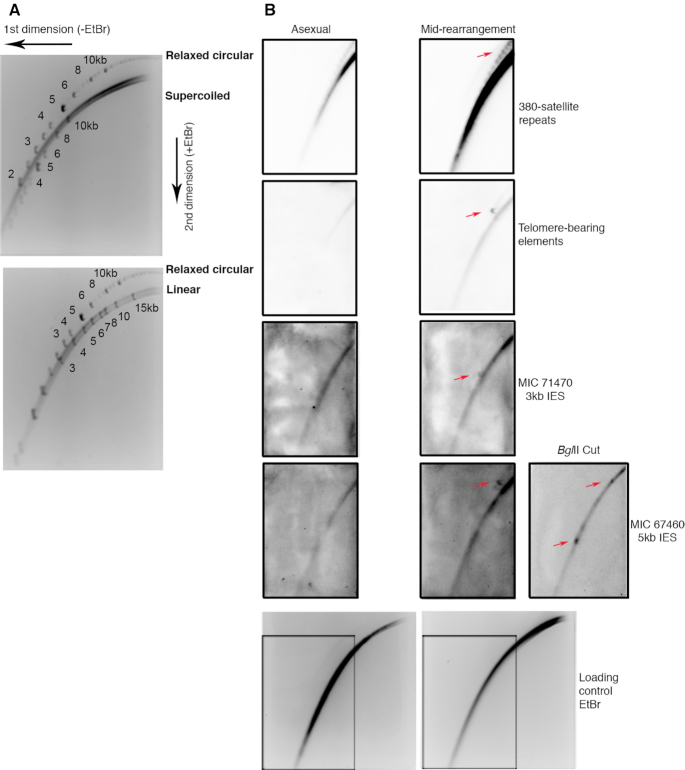
Figure 6.
Validating and further investigating the circular conformation and topology of bona fide eccDNA using 2D agarose gel electrophoresis. (A) Whole-cell DNA from asexually growing cells was separated on a 2D agarose gel with spiked-in relaxed circular, supercoiled circular and linear DNA standards showing separation of topologically different DNA molecules. Numbers represent size of the spike-in standards in kb. (B) Whole-cell DNA during asexual growth and rearrangement was separated on a 2D agarose gel and coupled to Southern blotting with probes specific for 380 nt satellite repeats, TBEs, a 5 kb and 3 kb IES that were annotated to contain high confidence circles. The same gel was stained with ethidium bromide to show equal loading of DNA. Membranes were stripped and reprobed sequentially for the four loci. Whole-cell DNA during mid-rearrangement was cut with BglII, which has a single target restriction site in the 5 kb IES circle in parental strain JRB310, leading to a 5 kb linear fragment. The other parental strain, JRB510, has two BglII restriction sites producing a 1.8 kb linear fragment that contains the probe hybridization target site. The cut DNA was separated on a 2D gel similar to the previous uncut samples and probed for the 5 kb IES circle. Red arrows mark the spots at the expected sizes for the two parental strains or overlapping the arc of linear DNA.