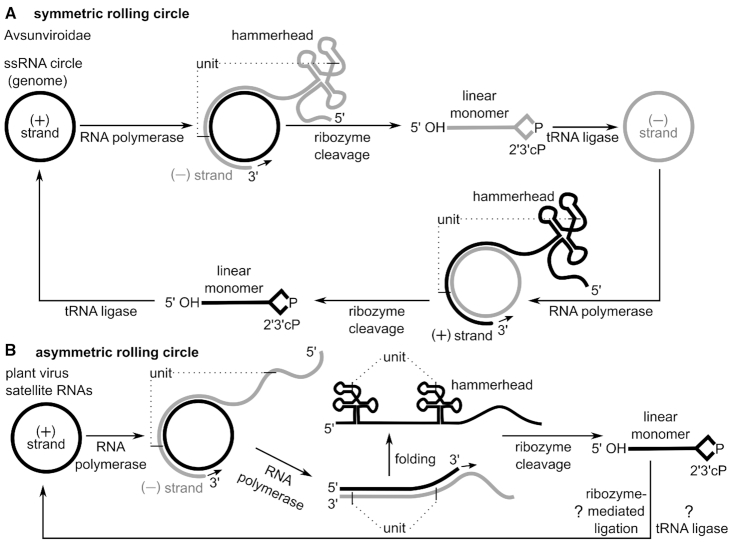
Figure 5.
Self-cleaving ribozymes support rolling circle replication mechanisms. (A) Symmetric rolling circle mechanism in viroids of the Avsunviroidae family and plant virus satellite RNAs. Circular (+) strand RNA is transcribed by DNA-dependent RNA polymerase into oligomeric (−) strand RNAs. Dotted lines indicate cleavage sites that define a single unit within the oligomeric RNA. Units are separated by hammerhead ribozyme cleavage and circularized by host enzymes, such as tRNA ligase. The circular (−) strand RNA is used for a second round of amplification yielding the (+) strand genome. In some plant virus satellite RNAs a hairpin instead of a hammerhead ribozyme could catalyse the cleavage of (+) strand oligomeric transcript, as well as potentially the ligation of the (+) strand linear monomer into a circular RNA. (B) Asymmetric rolling circle mechanism in plant virus satellite RNAs. First, the (+) strand RNA is transcribed by the host RNA polymerase into a long oligomeric transcript, the (−) strand RNA. Then the (−) strand RNA serves as template for a second transcription resulting in an oligomeric (+) strand. Hammerhead ribozymes, which are encoded in the (+) strand, cleave the oligomeric transcript into linear monomers. These unit-length transcripts are circularized either by ribozyme-mediated or enzymatic ligation.