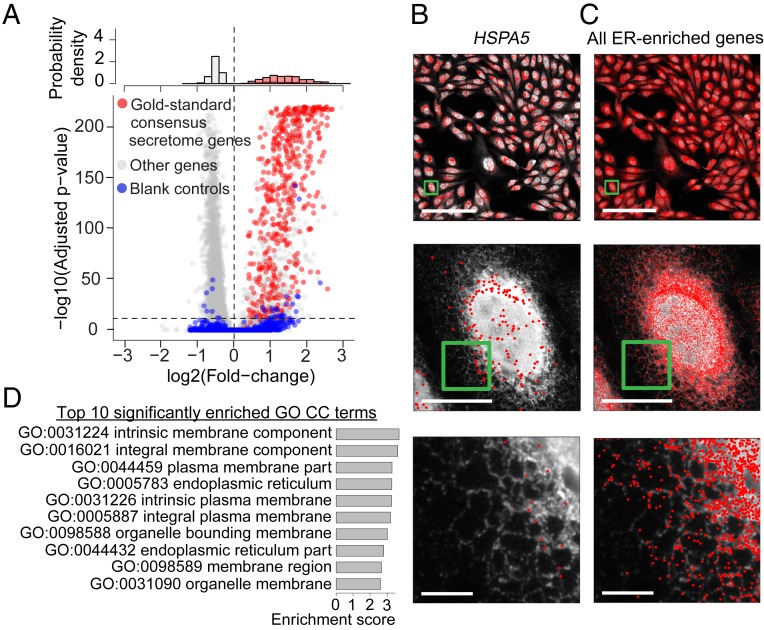
Fig. 2.
Identification of RNAs enriched at the endoplasmic reticulum. (A) Quantification of the ER enrichment for each RNA species. The fold change between count-per-million-normalized transcript counts localized to the ER versus those localized in the non-ER region of the cytoplasm and the corresponding P values were calculated for each gene. In cpm normalization, the abundance of each RNA species was divided by the abundance of all RNA species in the corresponding cellular compartment and multiplied by a million for each cell. P values are determined based on a 2-sided pairwise Wilcoxon rank-sum test across all cells and adjusted for multiple testing using Bonferroni correction. (A, Bottom) The scatterplot of the P value versus fold change for each gene. Gold-standard consensus secretome genes, other genes, and blank control barcodes are marked in red, gray, and blue, respectively. The horizontal dashed line indicates the P = 1e-10 significance threshold and the vertical dashed line indicates log2(fold change) = 0. (A, Top) Histograms of the fold changes for the gold-standard consensus secretome genes (red) and other genes (gray). For the other genes, only those with P < 1e-10 are shown in the histogram. (B) Spatial distribution of an example gene, HSPA5, that is enriched at the ER, overlaid on the ER image. (C) Spatial distribution of all genes identified as highly significantly enriched (log2[fold change] > 0, P < 1e-10) at the ER overlaid on the ER image. Each red point in B and C represents the position of a transcript detected by MERFISH from all 6 imaged z slices. The ER images in B and C are from 1 of the 6 imaged z slices. In B and C, Middle and Bottom panels are zoomed-in images of the boxed regions in the Top and Middle panels, respectively. (Scale bars: B and C, Top, 500 μm; B and C, Middle, 50 μm; B and C, Bottom, 10 μm.) (D) Top 10 significantly enriched (FDR < 0.05) GO cellular component terms among the genes highly significantly enriched at the ER (as described above), ordered by GO term enrichment score.