Significance
Dramatic and rapid changes in transcription take place upon heat shock (HS), where thousands of genes have been shown to be immediately up- or down-regulated in metazoans. The role of large-scale chromatin conformation and changes in long-range interactions between distal regulatory elements and HS-regulated promoters remains unclear. Our study shows that topologically associating domains and compartment structures remain remarkably unchanged upon acute HS in human and Drosophila, while only modest changes of distal regulatory interactions are observed in human cells. These results suggest that the global chromatin structure required for the HS response is preestablished across metazoans in order to drive transcriptional changes in HS responsive genes.
Keywords: heat shock, Hi-C, gene regulation, HSF1, TADs
Abstract
Heat shock (HS) initiates rapid, extensive, and evolutionarily conserved changes in transcription that are accompanied by chromatin decondensation and nucleosome loss at HS loci. Here we have employed in situ Hi-C to determine how heat stress affects long-range chromatin conformation in human and Drosophila cells. We found that compartments and topologically associating domains (TADs) remain unchanged by an acute HS. Knockdown of Heat Shock Factor 1 (HSF1), the master transcriptional regulator of the HS response, identified HSF1-dependent genes and revealed that up-regulation is often mediated by distal HSF1 bound enhancers. HSF1-dependent genes were usually found in the same TAD as the nearest HSF1 binding site. Although most interactions between HSF1 binding sites and target promoters were established in the nonheat shock (NHS) condition, a subset increased contact frequency following HS. Integrating information about HSF1 binding strength, RNA polymerase abundance at the HSF1 bound sites (putative enhancers), and contact frequency with a target promoter accurately predicted which up-regulated genes were direct targets of HSF1 during HS. Our results suggest that the chromatin conformation necessary for a robust HS response is preestablished in NHS cells of diverse metazoan species.
Proximal and distal regulatory elements coordinate cell type-specific transcriptional programs necessary for normal cellular function. Heat shock (HS) response is a well-studied model system for understanding gene regulation in metazoan organisms, including flies (1–5), mice (6), and humans (7, 8), causing both up-regulation of hundreds and down-regulation of thousands of target genes. HS induces binding of HSF1 to numerous heat shock elements (HSEs) across the genome. HSF1 binding to HSEs increases the rate at which paused RNA polymerase II (Pol II) is released into productive elongation at up-regulated genes (6, 8). Although the majority of HSF1-activated genes have promoter-bound HSF1, many do not (6, 7, 9). This indicates that HSF1 can regulate gene transcription through distant enhancer interactions.
The 3D structure of chromatin in the nucleus is proposed to play a fundamental role in gene regulation by facilitating or restricting regulatory element interactions. Gene activation during HS is linked to dramatic changes in chromatin. Loci encoding activated genes form highly visible puffs in polytene chromosomes of Drosophila upon HS (10, 11), and biochemical assays reveal massive changes in nuclease sensitivity and nucleosome loss in nonpolytene cells (12, 13). Dramatic transient changes in histone modification and chromatin composition also occur by the recruitment of specific transcription factors, chromatin remodelers, and histone modifiers (8, 14, 15). The extent of these changes along the chromosome and how they might influence long-range interactions between distal DNA sequences measured by Hi-C remains unclear.
In this study, we have used in situ Hi-C (16) to map the genomic contacts in human K562 and Drosophila S2 cells subjected to HS. We observed no evidence for global changes in compartments or topologically associating domains (TADs) in heat shocked cells, and only modest changes in contact frequency between HSF1 binding sites and their target genes. Despite the lack of changes in Hi-C data, integrating information about HSF1 binding strength and contact frequency with a target promoter accurately predicted which up-regulated genes were direct HSF1 targets. Thus, we propose that chromatin architecture necessary for HS response is preestablished in both human and Drosophila cells, potentially reflecting an evolutionarily conserved mechanism that enables cells to respond rapidly to stress.
Results
Global Chromatin Architecture Is Conserved During HS Despite Dramatic Transcriptional Changes.
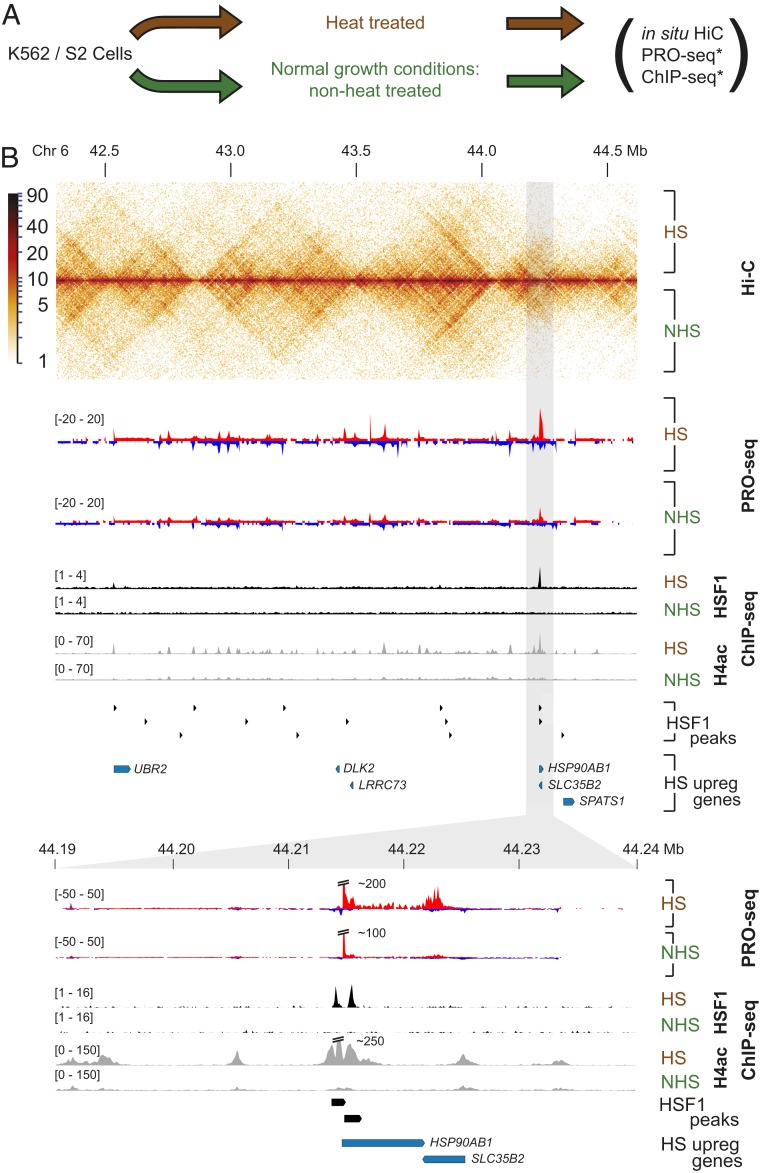
We have previously reported that HS induces transcriptional changes in thousands of genes in humans, mice, and Drosophila (5, 6, 8). To understand how changes in transcription correlate with changes in 3D chromatin architecture, we performed in situ Hi-C (16), both before and after 30 min of HS in the human chronic myelogenous leukemia K562 cell line (Fig. 1A). Hi-C libraries were sequenced to an estimated resolution of 10 kb in each condition (SI Appendix, Table S1). We confirmed that biological replicates were highly correlated (stratum-adjusted correlation coefficient [SCC] > 0.94) at 10-kb resolution using HiCRep (17), a method of correlating Hi-C data that compensates for distance dependence and domain structure (SI Appendix, Table S2). Comparison between HS and nonheat shock (NHS) Hi-C contact maps revealed a highly similar distribution of contact pairs across the genome (Fig. 1B). Genome-wide analysis using HiCRep revealed that heat maps from HS and NHS were correlated to the same extent as biological replicates (SI Appendix, Table S2). Thus, to a first approximation, we observed no evidence for differences in Hi-C contact maps between the 2 conditions.
Fig. 1.
Global chromatin architecture is unaltered during HS-induced transcriptional changes. (A) Schematic of datasets generated and analyzed in this study. Human K562 or Drosophila S2 HS and NHS cells were used to generate in situ Hi-C data, and these datasets were compared to corresponding PRO-seq (5, 8) and ChIP-seq (4, 7, 8) datasets denoted by asterisk. (B) Comparison of PRO-seq, in situ Hi-C, and ChIP-seq assays performed on HS and NHS K562 cells. Gray region highlights a classical HS locus containing the HSP90AB1 gene.
To determine whether HS changed chromatin conformation near HS-regulated genes, we first classified genes as HS up-regulated, down-regulated, or unregulated using PRO-seq data (8). As not all up-regulated genes depend on HSF1 (5, 6), we used PRO-seq data from control and HSF1 RNAi knockdown in K562 cells to classify up-regulated genes into HSF1-dependent or -independent categories (18). We classified 5,746 genes as unregulated with no detectable change in expression by PRO-seq, 227 genes as HSF1-dependent up-regulated, 360 as HSF1-independent up-regulated, and 4,002 genes as down-regulated (SI Appendix, Fig. S1).
Examination of Hi-C heatmaps near regions with HSF1 up- or down-regulated genes revealed similar patterns between HS and NHS. For instance, a closer examination of a locus harboring a classical HS gene, HSP90AB1, showed HS enriched HSF1 binding near the promoter, 4.3-fold transcriptional activation, and a dramatic increase in acetylation of histone 4. However, HS did not cause any obvious change in the composition of the Hi-C heatmaps (Fig. 1B). Thus, variation between NHS and HS conditions was relatively minor at the whole chromosome scale.
Chromosomal Compartmentalization Is Preserved upon HS.
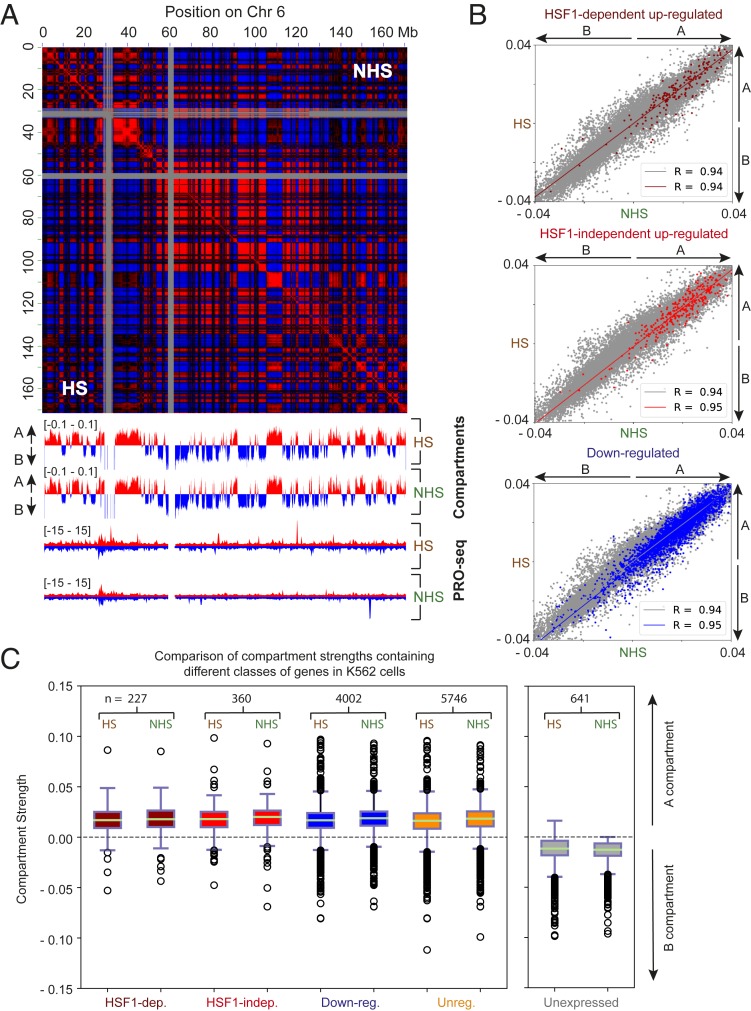
Hi-C has revealed the presence of active (A) and inactive (B) compartments, corresponding to regions of open, transcriptionally active chromatin, and closed, silent chromatin domains, respectively, in mammalian genomes (16, 19). A subset of compartments switch between active (A) and inactive (B) states in a manner that correlates with gene expression changes (20). To investigate whether short durations of HS change compartment organization, we identified compartments at 50-kb resolution in HS and NHS data. Examination of compartments in Juicebox (16) revealed that compartments were highly correlated between the NHS and HS conditions in K562 cells (Fig. 2A). Quantitative analysis of principal compartment scores revealed a high correlation in compartment strength genome-wide between NHS and HS conditions (Pearson’s R = 0.94).
Fig. 2.
Chromosomal compartmentalization is preserved upon HS. (A) Pearson’s correlation heatmap. Tracks below show first principal component (for both NHS and HS in K562 cells) used to call compartments (shown here for bin sizes of 50 kb). (B) Scatterplots show correlation between the strength of A and B compartment calls before and after HS. Gray, all compartments; dark red, compartments containing HSF1-dependent up-regulated genes (Top); red, compartments containing HSF1-independent up-regulated genes (Middle); blue, compartments containing down-regulated genes (Bottom). R = Pearson’s correlation coefficient. Solid line in each plot represents the best fit line (linear regression). (C) Boxplots showing the relative strength of compartments containing different classes of genes, for both HS and NHS conditions. Dark red, HSF1-dependent up-regulated genes; red, HSF1-independent up-regulated genes; blue, down-regulated genes; orange, unregulated genes; gray, unexpressed genes (separate boxplot showing only the relative strength of B compartments containing unexpressed genes in the NHS condition and the distribution of those same compartments after HS). n represents number of genes in each class.
If changes in transcription were accompanied by changes in chromatin compartmentalization, this would be most evident in compartments harboring HS-regulated genes. However, compartment calls for each category were highly correlated between the NHS and HS conditions with a Pearson’s correlation coefficient ranging from 0.94 to 0.95, suggesting no major difference in compartmentalization upon HS for any of these categories of genes (Fig. 2B). The small differences observed were similar in magnitude to those observed between biological replicates from the same HS conditions (R = 0.92 to 0.93) (SI Appendix, Fig. S2), suggesting that these differences are technical noise in the data rather than real biological signal. Both HS up- and down-regulated genes were predominantly localized in the active (A) compartment in both the NHS or HS conditions (Fig. 2C), consistent with previous observations of significant transcription of these genes before and after thermal stress (8). We observed small decreases in compartment strength for all classes of genes after HS. However, these changes were not correlated with the changes observed in transcription, indicating a transcription-independent mechanism. Taken together, these results show that compartmentalization remains largely unchanged during the first 30 min of HS.
Heat Shock Does Not Affect TAD Boundaries.
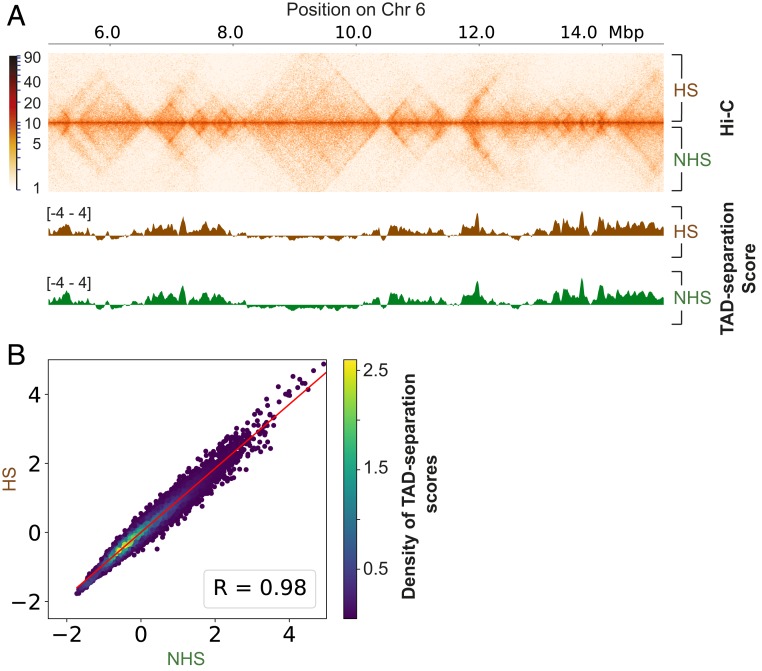
A previous study found evidence that TAD boundaries change following a short duration of heat stress in Drosophila (21). To determine whether TAD boundaries change in our Hi-C data, we computed the TAD-separation score (22) that allows measuring TAD boundary strength and comparing it between NHS and HS conditions. We found that TAD-separation scores were highly similar between HS and NHS conditions (Pearson’s R = 0.98), suggesting that TAD boundaries remain stable in response to 30 min of HS (Fig. 3 A and B and SI Appendix, Fig. S3).
Fig. 3.
Heat shock does not affect TAD boundaries. (A) Regions of 10 Mb of chromosome 6 (in K562) showing comparison of HS and NHS Hi-C contact matrices (Top). Comparison of TAD-separation scores for same region, for both the conditions (Bottom). (B) Correlation of TAD-separation scores for HS vs. NHS conditions, for all chromosomes in K562. R = Pearson’s correlation coefficient. Red line represents the best fit line (linear regression).
HSF1 Enhancer–Promoter Interactions Are Preestablished Prior to Heat Shock.
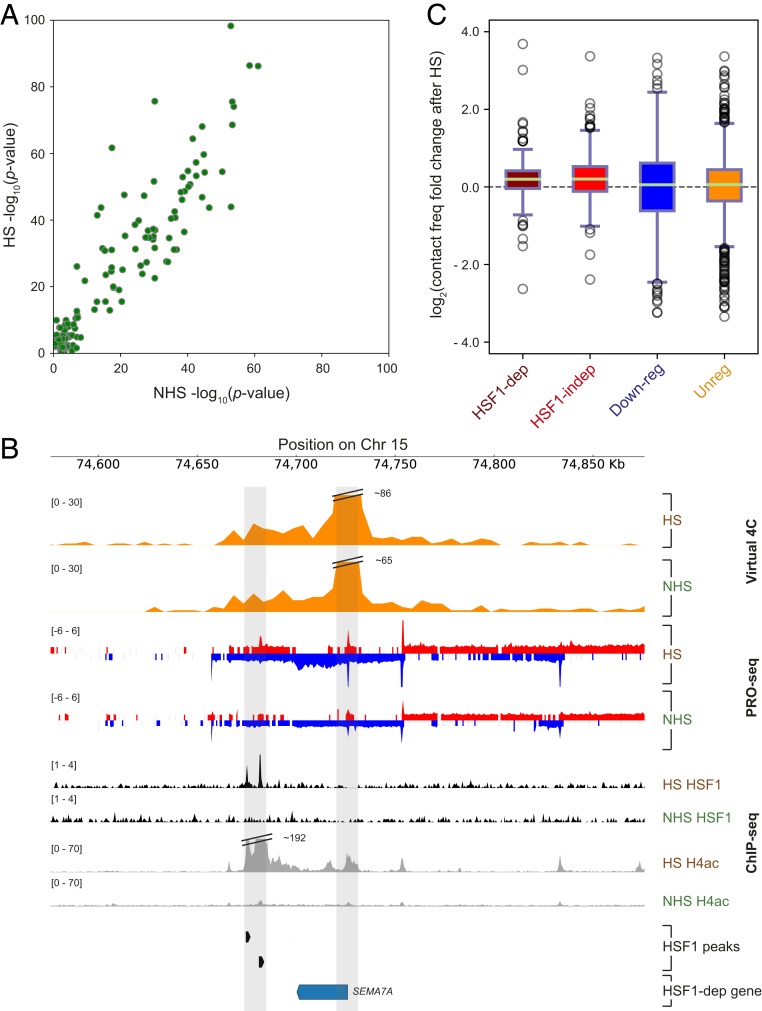
We asked whether interactions between promoter–enhancer pairs that are critical to establish the HS response were formed de novo following HS. We focused on HSF1-dependent up-regulated genes, for which the failure of the gene to activate following HSF1 knockdown provided functional evidence of an interaction between the gene and a proximal or distal HSF1 binding site. The majority of HSF1-dependent genes had an HSF1 binding site located within 1.5 kb of the transcription start site (TSS) (SI Appendix, Fig. S4). We devised a strategy to identify putative loop interactions between HSF1 binding sites and HSF1-dependent genes by comparing the contact frequency between HSF1 binding sites and HSF1-dependent genes with an empirical null distribution (SI Appendix, SI Methods). This strategy identified at least 1 HSF1 binding site significantly associated with 71% of HSF1-dependent genes at an empirical false positive ceiling of <10% (SI Appendix, SI Methods). This approach revealed substantial overlap between HS and NHS in loci with statistical evidence for interactions between HSF1 binding sites and HSF1-dependent up-regulated genes (126 out of 192; Fig. 4A and SI Appendix, Fig. S5A) that generally lie close to each other (median distance of 289 bp; SI Appendix, Fig. S5D). Additionally, cases where there was an apparent difference between HS and NHS generally had weaker statistical support where 1 condition was close to the cutoff threshold (SI Appendix, Fig. S5A). Thus, we conclude that the majority of HSF1-dependent genes are already proximal in 3D position to at least 1 HSF1 binding site in NHS cells.
Fig. 4.
Comparison of contact frequencies between HSF1-dependent gene promoters and HSF1 binding sites under NHS and HS conditions. (A) Significant looping interactions (P ≤ 0.01) between HSF1 binding sites and HSF1-dependent gene promoters for HS and NHS conditions (see also SI Appendix, Fig. S5A). (B) Virtual 4C plot showing contacts between SEMA7A TSS and its nearest HSF1 binding sites (gray bars). PRO-seq tracks are included to show changes in transcription for this gene upon HS. ChIP-seq tracks of HSF1 and H4ac show HSF1 binding and accompanying active chromatin status, respectively, upon HS. (C) Change in observed/expected looping interactions for HS and NHS Hi-C samples, for HSF1-dependent up-regulated, HSF1-independent up-regulated, down-regulated, and unregulated genes with HSF1 binding site, using a cutoff at which <10% of the unregulated interaction calls were significant.
Having observed no evidence for major qualitative differences in interacting enhancer–promoter pairs following HS, we asked whether HS induces quantitative differences in contact frequency between enhancers and their target promoters. We observed no evidence for a global difference in contact frequency between all pairs of HSF1-dependent promoters and the nearest distal (>10 kb) HSF1 binding site (SI Appendix, Fig. S6). For example, the HSF1-dependent up-regulated gene SEMA7A showed no evidence of changes in interactions upon HS with HSF1 binding sites located ∼50 kb downstream (Fig. 4B).
Recent studies found evidence of HS-dependent changes in contact frequency between interacting promoter–enhancer pairs (23). To test for more subtle differences, we examined contact frequencies between HSF1-dependent genes and HSF1 binding sites that showed statistical evidence for interactions in either HS or NHS (P < 0.01). Pairs of HSF1 binding sites and HSF1-dependent target genes were more likely to increase contact frequency in HS (P < 0.01, Wilcoxon rank sum test; Fig. 4C). We note that changes in contact frequency involving HSF1 binding sites were small and included both HSF1-dependent and HSF1-independent genes (median 20.5%; Fig. 4C). For example, a virtual 4C plot of the DGKE gene that enables visualizing contacts from DGKE as an anchor point shows a 27% increase in contact frequency between its TSS and its nearest HSF1 binding site after HS (SI Appendix, Fig. S5B). As a control, HS down-regulated and unregulated genes showed no difference in contact frequency with an interacting HSF1 binding site (Fig. 4C). We also analyzed contact frequency of the up-regulated distal transcriptional regulatory elements (dTREs) that show an increase in polymerase density upon HS, only a small fraction (5 to 10%) of which are bound by HSF1 (8). Interestingly, dTREs that gain polymerase density upon HS show small changes in contact frequency with both HSF1-dependent and HSF1-independent genes (SI Appendix, Fig. S5C).
Taken together, these results suggest that chromatin conformation necessary for response to HS is largely established in NHS conditions, but that a subset of enhancer–promoter interactions with strong statistical support do undergo small increases in Hi-C contact frequency following HS irrespective of HSF1 binding.
Chromatin Contacts Established before HS Accurately Predict HSF1-Dependent Genes.
A major unresolved problem in transcription regulation is identifying which enhancers regulate target genes. Having observed no substantial changes in enhancer–promoter interaction pairs, we asked whether the chromatin contacts necessary to facilitate a robust HS response were established in the NHS condition. Consistent with this hypothesis, simply the distance to the nearest HSF1 binding site predicted genes that were dependent on HSF1 with reasonably high accuracy (Fig. 5A and SI Appendix, Fig. S4). However, this criterion did not predict HSF1-dependent genes that were dependent on distal contacts, like SEMA7A.
Fig. 5.
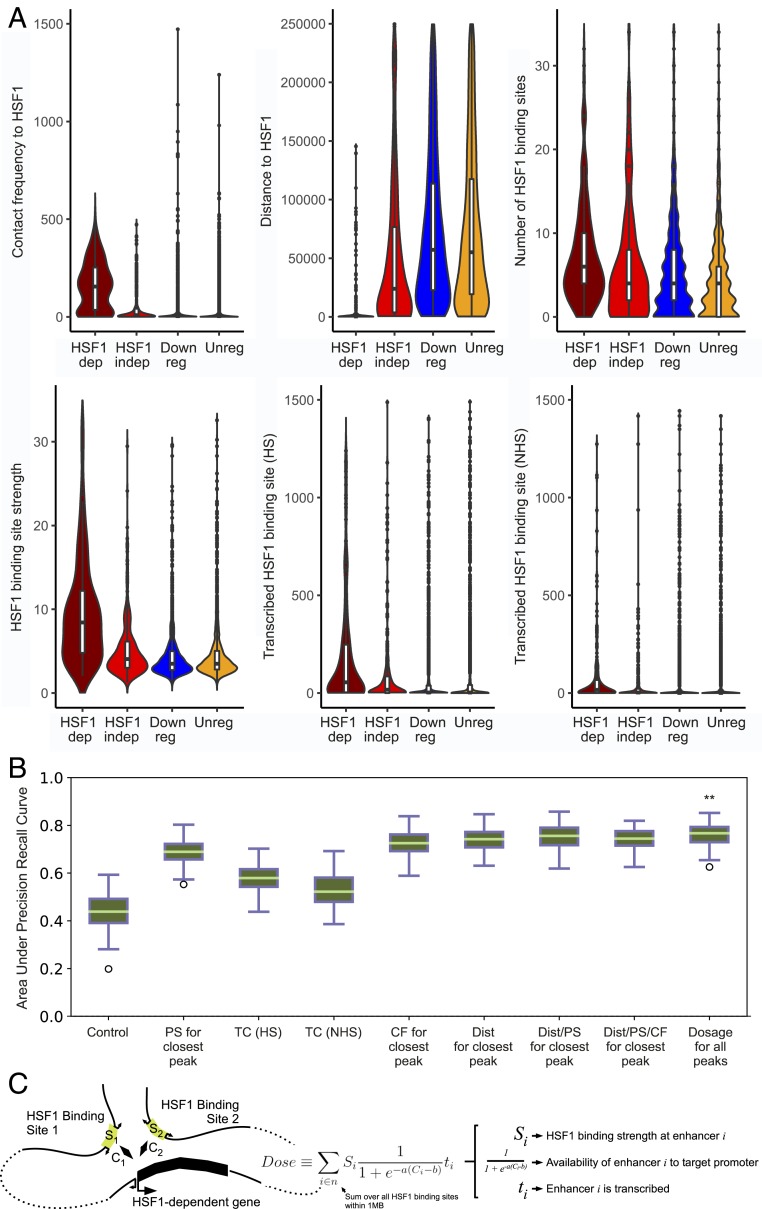
Prediction of HSF1-dependent up-regulated genes using NHS contact information. (A) Comparison of the correlation between HSF1-dependent gene activation and various genomic data. Panels show: the NHS contact frequency between genes’ TSS and the closest HSF1 binding site (Top Left); the linear distance between genes’ TSS and the closest HSF1 binding site (Top Middle); the number of HSF1 binding sites within a 2-Mb window around each gene TSS (Top Right); the strength (fold enrichment of HSF1 signal) of the closest HSF1 binding site (Bottom Left); transcription count (PRO-seq reads) at the closest HSF1 binding site for HS data (Bottom Middle); and for NHS data (Bottom Right). (B) Boxplots showing spread of area under the precision recall curve (auPRC) results for 1,000 iterations of various classifiers attempting to distinguish HSF1-dependent genes from HSF1-independent genes. From left to right, control: results obtained by randomly selecting the gene class; PS for closest peak: peak strength (fold enrichment of HSF1 binding signal) of the closest HSF1 binding site; TC (HS): transcription count (PRO-seq reads) at closest HSF1 binding site for HS data; TC (NHS): transcription count (PRO-seq reads) at closest HSF1 binding site for NHS data; CF for closest peak: contact frequency between genes’ TSS and the closest HSF1 binding site; Dist for closest peak: linear distance between genes’ TSS and the closest HSF1 binding site; Dist/PS for closest peak: linear distance/peak strength between genes’ TSS and the closest HSF1 binding site; Dist/PS/CF for closest peak: linear distance/peak strength/contact frequency between genes’ TSS and the closest HSF1 binding site; dosage for all peaks: scaled contact frequency multiplied by peak strength for all HSF1 binding sites within 1 Mb flanking each gene TSS. (C) Cartoon depicting components used to calculate HSF1 dosage.
Examination of the 49 HSF1-dependent genes that don’t have any detectable HSF1 binding within 10 kb of the TSS revealed that the majority of them still had a HSF1 binding site(s) located within the same TAD (n = 35/49; 71%).
We asked whether we could distinguish HSF1-dependent and -independent genes based on Hi-C contact frequencies, HSF1 binding location, and HSF1 binding strength. The number of HSF1 binding sites, the contact frequency between the HSF1 binding site and the promoter, HSF1 binding strength, and abundance of RNA polymerase at the HSF1 binding sites, were each correlated with whether genes were HSF1 dependent or not (Fig. 5A). Integrating these variables into a single classifier using gradient-boosted trees distinguished HSF1-dependent from HSF1-independent up-regulated genes much more accurately than random guessing, as determined by the area under the precision recall curve (auPRC) on holdout sites not used during model training (Fig. 5B and SI Appendix, Fig. S7). The best model used the distance between the promoter and the nearest HSF1 binding site, and the HSF1 binding strength, suggesting that simply distance and strength were enough to accurately classify most HSF1-dependent genes.
To develop a more biologically motivated classifier, we reasoned that HSF1 binding strength and the frequency of HSF1 binding site–promoter interactions were the 2 most important factors for a distal HSF1 binding site to regulate a target gene (24). We defined the “HSF1 dose” as the sum of all HSF1 binding sites within 1 Mb multiplied by their scaled contact frequency, and accounting for whether there is transcription at these HSF1 binding sites detectable by PRO-seq. (25). HSF1 dose improved the ability to predict HSF1 dependency of HS up-regulated genes slightly but significantly better than any other model (auPRC = 0.77; Fig. 5B). Notably, transcribed HSF1 binding sites had a larger effect on HSF1-dependent gene classification than nontranscribed enhancers, consistent with reports that many active enhancers are transcribed (26–28). Collectively, these results demonstrate that HSF1 dose (integrating HSF1 binding strength, transcription status, and contact frequency of nearby HSF1 binding sites) accurately predicted HSF1’s direct target genes.
Preprogrammed Chromatin Architecture Is Conserved Across Metazoans.
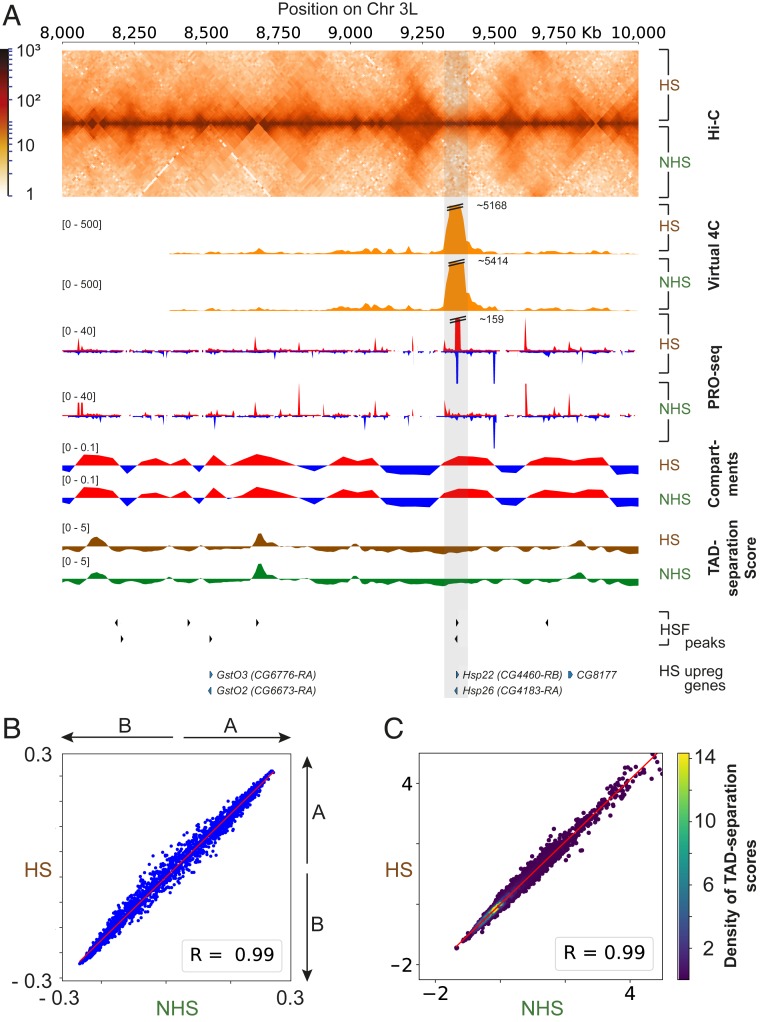
We asked whether chromatin architecture changes during heat stress in another metazoan organism. An earlier study with Drosophila Kc167 cells has shown that TAD structures undergo reorganization upon HS with a general reduction in border strength (21). However, our results with heat shocked Drosophila S2 cells revealed no significant changes in TAD structure, TAD-separation score, or compartmentalization, despite dramatic transcriptional activation in hundreds of genes (Fig. 6 A–C). Compartment calls and TAD-separation scores in NHS and HS conditions were found to be highly similar, with both having a Pearson’s coefficient of 0.99 (Fig. 6 B and C). The data recapitulated that observed in human K562 cells showing preestablished contacts between HSF-dependent up-regulated genes and their regulatory elements. We further analyzed the data published in NHS (29) and HS (21) conditions and found that technical variation between the NHS replicates could explain some of the differences in chromatin conformation reported in ref. 21 between NHS and HS cells (SI Appendix, Fig. S8). Our analysis suggests that response to HS in the context of 3D genome organization is prewired across metazoans, and this could be necessary to provide the “power” to rapidly drive the activation of genes having these preexisting connections.
Fig. 6.
Chromatin conformations are highly similar under NHS and HS conditions in Drosophila S2 cells. (A) Comparison of in situ Hi-C, virtual 4C, PRO-seq, compartmentalization, and TAD-separation scores for Drosophila S2 cells under NHS and HS conditions. (B) Correlation between the strength of compartment calls before and after HS. Plots were drawn as in Fig. 2B. (C) Correlation between TAD-separation scores before and after HS. Plots were drawn as in Fig. 3B.
Discussion
Chromosome conformation capture assays have provided powerful tools for interrogating chromatin contacts in specific cell types and conditions (30–34). Our understanding of the chromosome structure has been dramatically reshaped in recent years by the identification of TADs, sub-TADs, loops, compartments, and their roles in functional regulation of the genome in association with the architectural proteins (16, 19, 35–39). Generally, TADs are highly conserved across cell types, but they disappear along with compartments during mitosis (40). In differentiating cells, TADs are shown to be generally conserved, but disruption of their boundaries has been reported in cancer cells that could lead to oncogenesis (41). These studies indicate that in normal physiological circumstances, cells do not undergo significant rearrangements in TAD structure. However, there is evidence that some loop changes and compartmental switching occur during cellular differentiation and senescence associated with changes in gene expression (20, 42, 43).
Responses toward different environmental signals vary depending on the nature of the signal, cell type, and function. Transcriptional activation by HS is achieved by preconditioning the chromatin landscape of enhancers and promoters that allow establishment of promoter–proximal paused Pol II and the recruitment of critical transcription factors to release paused Pol II into productive elongation (8). In contrast, HS associated transcriptional repression of thousands of genes takes place by inhibiting paused Pol II release in mammals and reducing Pol II density along entire genes in flies (44). Our data suggest that HS does not alter TAD structures or weaken intra/inter-TAD boundaries in humans and flies. Additionally, we did not observe any significant switching or loss of compartmental strength following HS that is greater than that in our highly correlated biological replicates (SI Appendix, Fig. S2). Such an observation reemphasizes that the transcriptional response upon HS does not perturb global chromatin conformation; rather, the changes in paused Pol II densities at TSSs or across gene bodies are achieved primarily by loss or recruitment of transcription factors and chromatin remodelers (15, 44).
Our findings in the S2 cells contrast with the data and conclusions previously published for Drosophila Kc167 cells (21). This study reported a reduction in TAD border strength and increase in inter-TAD interactions accompanied by redistribution of architectural proteins upon a 20-min HS. This led to an interesting and surprising model where disrupted TAD boundaries following a thermal stress allow formation of Polycomb complex containing enhancer–promoter clusters that lead to gene repression. Such a dramatic reorganization model proposed in Kc167 cells seems inconsistent with the evolutionarily conserved transcriptional down-regulation seen in different ontological classes of genes across multiple species during HS (5, 6, 8). We did not analyze changes in Polycomb-mediated long-range interactions upon thermal stress, as it is beyond the scope of this study. However, if such interactions increase, they are not an effect of TAD reorganization, as our data show no detectable changes in TAD structures upon HS.
Cellular state and physiology appear to be critical in determining the dynamics of enhancer–promoter or promoter–promoter interactions. Although it has been shown that regulatory contacts are newly formed or strengthened while cells are undergoing transcriptional changes (45–47), there is also evidence of preestablished enhancer–promoter interactions during stimuli activation, differentiation, development, and stress (31, 45, 48, 49). This suggests that both dynamic and stable enhancer–promoter contacts could have contextual roles to regulate transcription of specific genes in a spatiotemporal manner. However, in the case of HS response, we observed that the vast majority of the contacts between HSF1-dependent genes and the HSF1 bound regulatory elements are preformed prior to HS. This result recapitulates the preestablished enhancer–promoter contacts as observed upon TNF-α stimulation of IMR90 cells (31) or during hypoxic stress in MCF-7 cells (49). Such evidence leads us to speculate that genomes have evolved to prewire not only the local chromatin architecture (8) but also the long-range regulatory interactions in 3D prior to stress so that the transcriptional response could be expedited. Cells are more frequently exposed to different kinds of stresses, including thermal, osmotic, hypoxic, and others compared to signaling cues for differentiation and development. It is possible that cells need an architectural platform to initiate immediate and rapid transcriptional response to survive thermal stress and a similar mechanism is likely to occur during other stresses as well. The similarity in the effects of heat shock on chromatin structure in humans and Drosophila suggests that the stress response mechanism is evolutionarily conserved in terms of regulatory interactions.
We used HSF1 as a model system to explore how transcription factors regulate target genes at a distance. Using data from an HSF1 knockdown, we identified transcriptional changes that were dependent on HSF1, but without HSF1 binding nearby the TSS. We found that Hi-C data can help to distinguish genes where up-regulation following HS was dependent on HSF1 from those where it was not. The best model integrated the binding strength of HSF1 (as determined by ChIP-seq), the presence of eRNA transcription at an HSF1 binding site, and the Hi-C contact frequency. Our observations are consistent with the notion that Hi-C data are a surrogate for the degree to which a distal enhancer is in a position where it is able to regulate target promoters (although we note that simply the distance between the HSF1-dependent TSS and the nearest HSF1 binding site performed nearly as well in our test). Furthermore, data from the NHS condition was just as informative as data from HS. Taken together, our results suggest that chromatin contacts observed in Hi-C which are necessary for a robust HS response are all in place prior to thermal stress.
Our study investigates an important connection of transcriptional regulation to chromatin interactions. In summary, we find that chromatin interactions between regulatory elements and their target promoters appear to be mostly prewired, and that the massive changes in transcription and chromatin following HS are not accompanied by significant changes in TADs, TAD boundaries, and compartments. Although we do not see major changes in these chromatin conformations after a 30-min HS, when transcription has already changed dramatically at genes and enhancers, we find a subset of HS up-regulated genes that show a modest increase in contact frequency upon HS. Surprisingly this change is not restricted to HSF1-dependent genes but also, include the HSF1-independent genes. Chromatin decondensation caused by HS could explain the slight increase in interactions between HS up-regulated genes and the up-regulated dTREs. Further perturbation studies involving these enhancers and promoters along with development of higher resolution assays could allow delving more deeply into changes in chromatin architecture and interactions triggered by the stress response.
Materials and Methods
Hi-C.
Human K562 and Drosophila S2 cells were subjected to HS or not (NHS) and cross-linked with 1% formaldehyde for 10 min at room temperature followed by quenching with glycine for 5 min. In situ Hi-C was performed based on the protocol described previously (16). Further details are provided in SI Appendix, Materials and Methods.
Hi-C Data Analysis.
Files containing sequenced read pairs were processed using the Juicer pipeline as described previously (16). Reads for human K562 cells and Drosophila S2 cells were aligned to hg19 and dm3, respectively. We required that all alignments were high quality by filtering for a MAPQ score greater than 30.
Reads for Drosophila Kc167 cells (SI Appendix, Fig. S8) from refs. 21 and 29 (available at GEO database accessions GSE63518 and GSM942889, respectively) were processed in a similar fashion (i.e., aligned to dm3 and processed using the Juicer pipeline).
Map resolution was calculated according to the definition proposed in ref. 16, as the smallest bin size such that 80% of loci have at least 1,000 contacts. For human K562 data, we used scripts that were part of the Juicer pipeline to compute resolution (50). For Drosophila, we used an alternative script specifically designed for the Drosophila genome (51), because we noted inconsistent results using Juicer. We used this definition to determine the finest scale at which one can reliably discern local features. Details of computational methods and analyses are provided in SI Appendix, Materials and Methods.
Additional Datasets Used in This Study.
The following datasets were also used in this study: K562 HSF1 ChIP-seq data, GSE43579 (7); K562 PRO-seq data, GSE89230 (8); K562 H4ac ChIP-seq data, GSE89382 (8); S2 HSF ChIP-seq data, GSE19025 (4); and S2 PRO-seq data, GSE77607 (5).
Code Repository.
All other code was custom written in Python 2.7. The significant parts of this code, and example data, are available on the Danko Lab’s GitHub website (https://github.com/Danko-Lab/HS_transcription_regulation).
Danko-Lab/Hi-C_contact_caller is the program for determining the significance of interactions for pairs of points within a chromosome using a Hi-C contact map (https://github.com/Danko-Lab/Hi-C_contact_caller, version: 88efbbf).
Supplementary Material
Acknowledgments
We thank members of the Lis and Danko labs for thoughtful discussions about this work. C.G.D., A.O., and J.T.L. acknowledge support from the National Institutes of Health Common Fund 4D Nucleome Program (Grant U01HL129958). Additional support was provided by National Human Genome Research Institute R01-HG009309 and an Nvidia GPU grant (to C.G.D.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
Data deposition: The Hi-C data have been deposited at the Gene Expression Omnibus (GEO) database, (accession no. GSE130778). Custom code and example data have been deposited in GitHub, https://github.com/Danko-Lab/HS_transcription_regulation. The Hi-C contact caller program has been deposited in GitHub, https://github.com/Danko-Lab/Hi-C_contact_caller (version: 88efbbf).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1901244116/-/DCSupplemental.
References
- 1.Ritossa F., A new puffing pattern induced by temperature shock and DNP in drosophila. Experientia 18, 571–573 (1962). [Google Scholar]
- 2.Yao J., Munson K. M., Webb W. W., Lis J. T., Dynamics of heat shock factor association with native gene loci in living cells. Nature 442, 1050–1053 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Guertin M. J., Petesch S. J., Zobeck K. L., Min I. M., Lis J. T., Drosophila heat shock system as a general model to investigate transcriptional regulation. Cold Spring Harb. Symp. Quant. Biol. 75, 1–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guertin M. J., Lis J. T., Chromatin landscape dictates HSF binding to target DNA elements. PLoS Genet. 6, e1001114 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duarte F. M., et al. , Transcription factors GAF and HSF act at distinct regulatory steps to modulate stress-induced gene activation. Genes Dev. 30, 1731–1746 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mahat D. B., Salamanca H. H., Duarte F. M., Danko C. G., Lis J. T., Mammalian heat shock response and mechanisms underlying its genome-wide transcriptional regulation. Mol. Cell 62, 63–78 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vihervaara A., et al. , Transcriptional response to stress in the dynamic chromatin environment of cycling and mitotic cells. Proc. Natl. Acad. Sci. U.S.A. 110, E3388–E3397 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vihervaara A., et al. , Transcriptional response to stress is pre-wired by promoter and enhancer architecture. Nat. Commun. 8, 255 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hahn J. S., Hu Z., Thiele D. J., Iyer V. R., Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol. Cell. Biol. 24, 5249–5256 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashburner M., Patterns of puffing activity in the salivary gland chromosomes of Drosophila. V. Responses to environmental treatments. Chromosoma 31, 356–376 (1970). [DOI] [PubMed] [Google Scholar]
- 11.Boehm A. K., Saunders A., Werner J., Lis J. T., Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol. Cell. Biol. 23, 7628–7637 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petesch S. J., Lis J. T., Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell 134, 74–84 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu C., Wong Y. C., Elgin S. C., The chromatin structure of specific genes: II. Disruption of chromatin structure during gene activity. Cell 16, 807–814 (1979). [DOI] [PubMed] [Google Scholar]
- 14.Saunders A., et al. , Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo. Science 301, 1094–1096 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Petesch S. J., Lis J. T., Overcoming the nucleosome barrier during transcript elongation. Trends Genet. 28, 285–294 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rao S. S., et al. , A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang T., et al. , HiCRep: Assessing the reproducibility of Hi-C data using a stratum-adjusted correlation coefficient. Genome Res. 27, 1939–1949 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vihervaara A., et al. , Stress-induced transcriptional memory accelerates promoter-proximal pause-release and decelerates termination over mitotic divisions. bioRxiv:10.1101/576959 (14 March 2019). [DOI] [PMC free article] [PubMed]
- 19.Lieberman-Aiden E., et al. , Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dixon J. R., et al. , Chromatin architecture reorganization during stem cell differentiation. Nature 518, 331–336 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li L., et al. , Widespread rearrangement of 3D chromatin organization underlies polycomb-mediated stress-induced silencing. Mol. Cell 58, 216–231 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramírez F., et al. , High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat. Commun. 9, 189 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyu X., Rowley M. J., Corces V. G., Architectural proteins and pluripotency factors cooperate to orchestrate the transcriptional response of hESCs to temperature stress. Mol. Cell 71, 940–955.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fulco C. P., et al. , Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science 354, 769–773 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Danko C. G., et al. , Identification of active transcriptional regulatory elements from GRO-seq data. Nat. Methods 12, 433–438 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henriques T., et al. , Widespread transcriptional pausing and elongation control at enhancers. Genes Dev. 32, 26–41 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mikhaylichenko O., et al. , The degree of enhancer or promoter activity is reflected by the levels and directionality of eRNA transcription. Genes Dev. 32, 42–57 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Core L. J., et al. , Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat. Genet. 46, 1311–1320 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hou C., Li L., Qin Z. S., Corces V. G., Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol. Cell 48, 471–484 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanyal A., Lajoie B. R., Jain G., Dekker J., The long-range interaction landscape of gene promoters. Nature 489, 109–113 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin F., et al. , A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature 503, 290–294 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mifsud B., et al. , Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat. Genet. 47, 598–606 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Li G., et al. , Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148, 84–98 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmitt A. D., et al. , A compendium of chromatin contact maps reveals spatially active regions in the human genome. Cell Rep. 17, 2042–2059 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dixon J. R., et al. , Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanborn A. L., et al. , Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl. Acad. Sci. U.S.A. 112, E6456–E6465 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rao S. S. P., et al. , Cohesin loss eliminates all loop domains. Cell 171, 305–320.e24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nora E. P., et al. , Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169, 930–944.e22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vian L., et al. , The energetics and physiological impact of cohesin extrusion. Cell 173, 1165–1178.e20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Naumova N., et al. , Organization of the mitotic chromosome. Science 342, 948–953 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valton A. L., Dekker J., TAD disruption as oncogenic driver. Curr. Opin. Genet. Dev. 36, 34–40 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Criscione S. W., et al. , Reorganization of chromosome architecture in replicative cellular senescence. Sci. Adv. 2, e1500882 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siersbaek R., et al. , Dynamic rewiring of promoter-anchored chromatin loops during adipocyte differentiation. Mol. Cell 66, 420–435.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Vihervaara A., Duarte F. M., Lis J. T., Molecular mechanisms driving transcriptional stress responses. Nat. Rev. Genet. 19, 385–397 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rubin A. J., et al. , Lineage-specific dynamic and pre-established enhancer-promoter contacts cooperate in terminal differentiation. Nat. Genet. 49, 1522–1528 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li W., et al. , Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 498, 516–520 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tolhuis B., Palstra R. J., Splinter E., Grosveld F., de Laat W., Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell 10, 1453–1465 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Ghavi-Helm Y., et al. , Enhancer loops appear stable during development and are associated with paused polymerase. Nature 512, 96–100 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Platt J. L., et al. , Capture-C reveals preformed chromatin interactions between HIF-binding sites and distant promoters. EMBO Rep. 17, 1410–1421 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Durand N. C., et al. , Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 3, 95–98 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eagen K. P., Aiden E. L., Kornberg R. D., Polycomb-mediated chromatin loops revealed by a subkilobase-resolution chromatin interaction map. Proc. Natl. Acad. Sci. U.S.A. 114, 8764–8769 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.