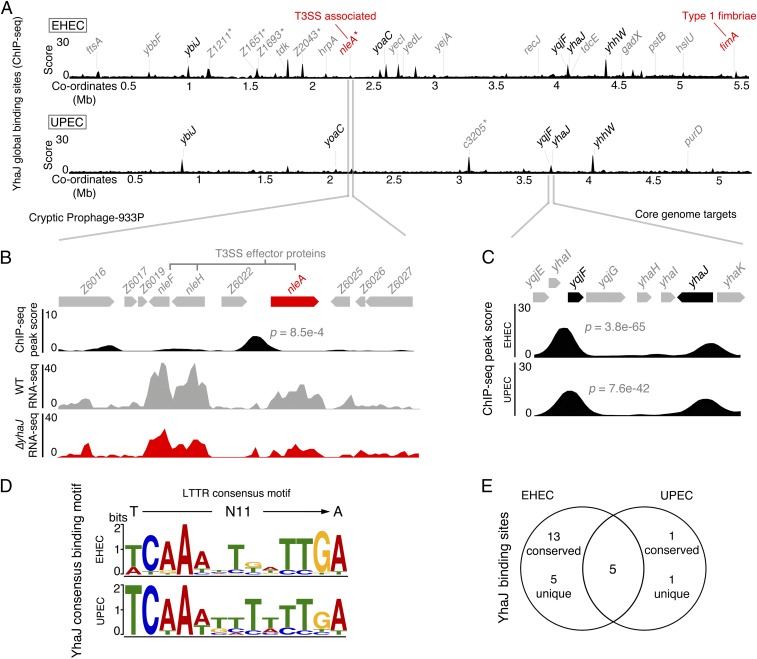
Fig. 2.
YhaJ binds to unique core and horizontal chromosomal locations in distinct E. coli pathotypes. (A) Global chromosome binding dynamics of YhaJ in EHEC and UPEC identified by ChIP-seq. Significant peaks (P ≤ 0.01; 2 biological replicates) related to known targets are labeled in black, novel targets are in gray, and pathotype-specific targets are marked with an asterisk. The virulence-associated targets nleA (T3SS-associated) and fimA (type 1 fimbriae) are highlighted in red. (B) Expanded view of the horizontally acquired CP-933P locus in EHEC. The prophage region encodes multiple non–LEE-encoded effectors as indicated. The ChIP-seq track illustrates the identified YhaJ binding site upstream of nleA and RNA-seq data illustrate the downshift in transcription from nleA for ΔyhaJ (red) versus WT EHEC (gray). (C) Expanded view of ChIP-seq peaks identified for 2 common core genome targets, yhaJ itself and yqjF. The P values for yqjF peaks are indicated. (D) The computed consensus binding motif for YhaJ targets in EHEC and UPEC based on ChIP-seq data. The motifs match the generalized consensus sequence for LTTRs (T-N11-A) as illustrated above. (E) Comparison of YhaJ binding sites in EHEC and UPEC. ChIP-seq experiments were performed in biological duplicate.