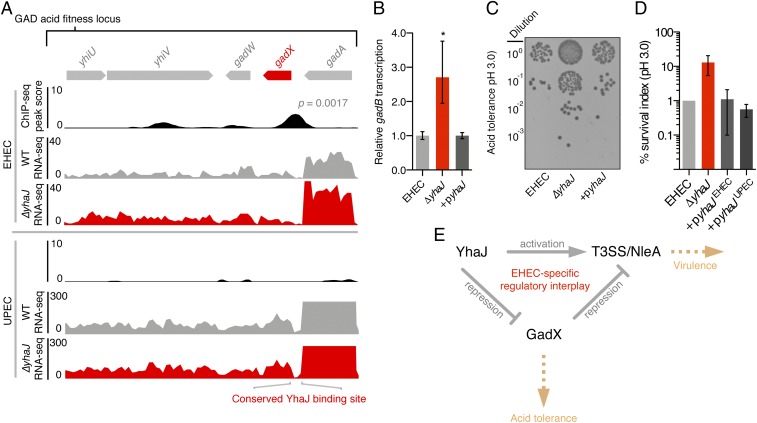
Fig. 3.
YhaJ overrides the regulation of EHEC acid tolerance to control virulence via direct and indirect routes. (A) Expanded view of the EHEC YhaJ binding site upstream of the GAD acid tolerance regulator gene gadX. (A, Upper) RNA-seq tracks show the upshift in GAD gene transcription for ΔyhaJ (red) versus WT EHEC (gray). (A, Lower) Tracks reveal the lack of this regulation in a UPEC genetic background. The conserved sequence of the YhaJ-binding motif in both strains is illustrated (Bottom). (B) Repression of acid tolerance by YhaJ confirmed by qRT-PCR analysis of the GadX-regulated gene gadB in WT EHEC, ΔyhaJ, and +pyhaJ backgrounds. *P ≤ 0.05 derived from 3 biological replicates ±SD (Student’s t test). (C) Acid tolerance assay of WT EHEC, ΔyhaJ, and complemented (+pyhaJ) cells exposed to acidic conditions (pH 3.0). (D) Quantification of CFUs from B representing the mean survival relative to the WT (±SD of 4 biological replicates). The EHEC ΔyhaJ phenotype was complemented with both the EHEC and UPEC yhaJ alleles expressed in trans (+pyhaJEHEC and +pyhaJUPEC, respectively). (E) The regulatory interplay between YhaJ and GadX in fine-tuning T3SS expression in EHEC. Pointed and blunt arrows represent activation and repression, respectively, whereas the yellow arrows indicate the associated phenotype of each pathway.