Abstract
Wilms tumor is the most common type of renal malignancy in children. Previous studies have demonstrated that single nucleotide polymorphisms (SNPs) in the AURKA gene could predispose to several human malignancies. We recruited 145 cases and 531 cancer-free controls to investigate whether AURKA gene variants modify Wilms tumor susceptibility. Three AURKA SNPs (rs1047972 C>T, rs2273535 T>A, and rs8173 G>C) were genotyped by the Taqman methodology. Odds ratios (ORs) and 95% confidence intervals (CIs) were used to assess the strength of association between AURKA SNPs and Wilms tumor risk. We found that only the rs8173 G>C polymorphism was significantly associated with Wilms tumor risk (GC vs. GG: adjusted OR (AOR) = 0.50, 95% CI = 0.35–0.73, P=0.0002; GC/CC vs. GG: AOR = 0.60, 95% CI = 0.42–0.88, P=0.008). Stratification analysis revealed that rs8173 GC/CC genotypes were associated with Wilms tumor risk among children aged >18 months (AOR = 0.56, 95% CI = 0.34–0.93, P=0.024), male children (AOR = 0.54, 95% CI = 0.33–0.90, P=0.017), and children with clinical stage III + IV diseases (AOR = 0.56, 95% CI = 0.35–0.90, P=0.017). Haplotype analysis indicated that the CAG haplotype was significantly associated with increased Wilms tumor risk. In conclusion, our findings indicated that the AURKA rs8173 G>C polymorphism was associated with decreased Wilms tumor risk in Chinese children.
1. Introduction
Wilms tumor (WT), also known as nephroblastoma, is the most common renal malignancy in children [1, 2]. It accounts for 6% to 7% of malignant tumors in children under the age of 15 years, with an incidence rate of about 7–10 cases per million in Western countries [3]. The incidence rate of WT in Chinese is around 3.3 cases per million [4]. Over the past 20 years, because of the application of multimodality treatment, including surgery, radiotherapy, chemotherapy, and autologous stem cell transplantation, the overall survival rate of 5 years has increased from 30% to 90% [2, 5–7]. Despite the great achievements in the treatment of WT, the prognosis of nearly 25% of patients with high-risk diseases remains unsatisfying [8]. In addition, about 25% of survivors suffer from the high cost of treatment and the physical torment of some chronic conditions [9, 10].
Previous studies have found that genetic factors contribute to the risk of Wilms tumor and identified a number of genes associated with WT, including Wilms' tumor protein 1 (WT1), β-catenin, tumor protein 53 (TP53), catenin beta 1 (CTNNB1), and AMER1 [11–13]. Although recent genome-wide studies on WT have revealed some previously unknown gene mutations implicated with WT, current known genetic variants are not adequate to fully elucidate the pathogenesis of WT [14]. Therefore, it is particularly important to identify more causal genetic variants for WT.
Aurora kinase A (AURKA) is located at chromosome 20q13.2, consisting of 12 exons [15]. It encodes a centrosome-related serine/threonine kinase found to be overexpressed in many human cancers, including primary colorectal carcinoma, esophageal squamous cell carcinoma, neuroblastoma, and breast and ovarian cancers [16–19]. Overexpression of AURKA leads to abnormal centrosome amplification, thereby affecting the stability of the genome and inducing tumorigenesis [16, 20]. Furthermore, inhibition of AURKA expression could result in abnormal cell mitosis, eventually leading to cell death [21–23]. Therefore, AURKA might be an attractive target for anticancer therapies [24].
In addition, previous studies had found that the AURKA gene single nucleotide polymorphism (SNP) was closely related to the risk of several human malignancies [25–27]. Early research demonstrated that the AA genotype of AURKA rs2273535 T>A could increase the risk of oral cancer [28]. Studies in recent years have also found that AURKA rs2273535 T>A polymorphism could enhance the susceptibility of gastric cancer [15]. Furthermore, there were related studies reported that Caucasians carrying AURKA rs1047972 T>C had a relatively low risk of developing breast cancer [26]. However, studies regarding their association with WT risk are lacking. Therefore, we performed this case-control study with 145 cases of WT and 531 control subjects to assess the association between AURKA gene polymorphisms and WT susceptibility in Chinese children.
2. Materials and Methods
2.1. Study Subjects
In this study, a total of 145 cases of WT and 531 controls were collected from the Guangzhou Women and Children Medical Center from 2001 to 2016, as we described previously [29]. Controls were selected from those receiving routine physical examination during the same period. The cases included in this study were all sporadic and the control groups were confirmed free of history of malignancy. Both WT patients and controls were frequency-matched based on the age, gender, and ethnicity. The frequency distribution information of selected variables for WT cases and cancer-free controls was summarized in Supplementary . We obtained written informed consents from the parents or legal guardians of all subjects. The study protocol was approved by the Institutional Review Board of the Guangzhou Women and Children's Center (Ethical Approval Number: 2018022102).
2.2. SNP Selection and Genotyping
The selection criteria for potentially functional polymorphisms have been described in detail previously [30]. Briefly, we choose potential functional polymorphisms in the 5′-flanking region, 5′UTR, 3′UTR, and exons of the AURKA gene. Three SNPs in the AURKA gene were finally selected (rs1047972 C>T (Assay ID: AN322ZF), rs2273535 T>A (Assay ID: C__25623289_10), and rs8173 G>C (Assay ID: C__8947675_10)). The rs1047972 C>T polymorphism may lead to amino acid alteration from Val to Ile at codon 57; the rs2273535 T>A polymorphism may lead to splicing alteration, whereas the rs8173 G>C polymorphism may lead to miRNA binding alteration. These three SNPs could capture additional fourteen SNPs (Supplementary ). Moreover, there was no significant linkage disequilibrium between paired polymorphisms in Supplementary (rs8173/rs1047972, R2 = 0.119; rs8173/rs2273535, R2 = 0.527; rs1047972/rs2273535, R2 = 0.291) [31]. Human genomic DNA was mainly extracted from the peripheral blood samples using the TIANamp Blood DNA Kit (TianGen Biotech, Beijing, China). The mean yield of DNA was 12.21 μg (median = 6.25 μg; range = 1.08 to 60 μg), with a mean A260/A280 ratio of 1.89 (median = 1.90; range = 1.78 to 2.05). AURKA SNPs were genotyped using the ABI-7900 Sequence Detection System (Applied Biosystem, Foster City, CA) and Taqman real-time PCR, as described previously [30]. The information on all the genotyped samples for all three SNPs was summarized in Supplementary .
2.3. Genotype and Gene Expression Correlation Analysis
The FAM210B mRNA expression data were retrieved for transformed fibroblasts cells with different genotypes of AURKA SNPs from the GTEx Portal database (https://www.gtexportal.org/home/). These data were used to assess the correlation between rs8173 G>C genotypes and mRNA expression-level alteration [32].
2.4. Statistical Analysis
All statistical analysis of data was processed by SAS (version 9.4; SAS Institute, Cary, NC, USA). The distribution of subject characteristics between cases and controls was examined by a bilateral χ2 test. The goodness-of-chi-squared test was used to validate whether the genotype frequencies in the controls were consistent with the Hardy–Weinberg equilibrium (HWE). Multivariate logistic regression analysis was adopted to calculate the odds ratios (ORs) and 95% confidence intervals (CIs). The adjusted ORs and 95% CIs were calculated using an unconditional logistic regression model with adjustment for age and gender to assess the association between AURKA polymorphisms and WT risk. Inferred haplotypes of AURKA gene were based on observed genotypes, and adjusted ORs and 95% CIs were obtained by logistic regression models with adjustment for age and gender. P < 0.05 was considered to be statistically significant.
3. Results
3.1. Association between AURKA Gene Polymorphisms and Wilms Tumor Susceptibility
In this study, 143 cases and 531 controls were successfully genotyped. All selected SNP genotypes had a frequency distribution consistent with HWE (rs1047972 C>T, P=0.598; rs2273535 T>A, P=0.701; rs8173 G>C, P=0.272). The genotype frequencies of the case group and control group are shown in Table 1. We found rs8173 G>C polymorphism was significantly associated with decreased WT risk (GC vs. GG: adjusted OR (AOR) = 0.50, 95% CI = 0.35–0.73, P=0.0002; GC/CC vs. GG: AOR = 0.60, 95% CI = 0.42–0.88, P=0.008).
Table 1.
Association between AURKA gene polymorphisms and Wilms tumor susceptibility.
| Genotype | Cases (N = 143) | Controls (N = 531) | P a | Crude OR (95% CI) | P | Adjusted OR (95% CI)b | P b |
|---|---|---|---|---|---|---|---|
| rs1047972 C>T (HWE = 0.598) | |||||||
| CC | 110 (76.92) | 412 (77.59) | 1.00 | 1.00 | |||
| CT | 30 (20.98) | 110 (20.72) | 0.89 (0.57–1.39) | 0.613 | 0.89 (0.57–1.39) | 0.622 | |
| TT | 3 (2.10) | 9 (1.69) | 1.09 (0.29–4.08) | 0.898 | 1.10 (0.29–4.10) | 0.891 | |
| Additive | 0.945 | 1.05 (0.71–1.55) | 0.809 | 1.04 (0.71–1.54) | 0.834 | ||
| Dominant | 33 (23.08) | 119 (22.41) | 0.866 | 1.04 (0.67–1.61) | 0.865 | 1.03 (0.66–1.60) | 0.889 |
| Recessive | 140 (97.90) | 522 (98.31) | 0.746 | 1.24 (0.33–4.65) | 0.747 | 1.22 (0.33–4.59) | 0.764 |
|
| |||||||
| rs2273535 T>A (HWE = 0.701) | |||||||
| TT | 66 (46.15) | 234 (44.07) | 1.00 | 1.00 | |||
| TA | 65 (45.45) | 234 (44.07) | 0.82 (0.57–1.16) | 0.263 | 0.82 (0.57–1.17) | 0.265 | |
| AA | 12 (8.39) | 63 (11.86) | 0.56 (0.29–1.08) | 0.084 | 0.56 (0.29–1.08) | 0.085 | |
| Additive | 0.502 | 0.88 (0.67–1.17) | 0.377 | 0.88 (0.67–1.17) | 0.374 | ||
| Dominant | 77 (53.85) | 297 (55.93) | 0.656 | 0.92 (0.63–1.33) | 0.656 | 0.92 (0.63–1.33) | 0.659 |
| Recessive | 131 (91.61) | 468 (88.14) | 0.241 | 0.68 (0.36–1.30) | 0.244 | 0.68 (0.35–1.29) | 0.237 |
|
| |||||||
| rs8173 G>C (HWE = 0.272) | |||||||
| GG | 71 (49.65) | 196 (36.91) | 1.00 | 1.00 | |||
| GC | 54 (37.76) | 263 (49.53) | 0.51 (0.35–0.73) | 0.0003 | 0.50 (0.35–0.73) | 0.0002 | |
| CC | 18 (12.59) | 72 (13.56) | 0.62 (0.35–1.08) | 0.090 | 0.61 (0.35–1.08) | 0.088 | |
| Additive | 0.018 | 0.74 (0.56–0.98) | 0.033 | 0.75 (0.56–0.99) | 0.040 | ||
| Dominant | 72 (50.35) | 335 (63.09) | 0.006 | 0.59 (0.41–0.86) | 0.006 | 0.60 (0.42–0.88) | 0.008 |
| Recessive | 125 (87.41) | 459 (86.44) | 0.762 | 0.92 (0.53–1.60) | 0.762 | 0.92 (0.53–1.61) | 0.781 |
OR, odds ratio; CI, confidence interval; HWE, Hardy–Weinberg equilibrium. aχ2 test for genotype distributions between Wilms tumor patients and controls; badjusted for age and gender.
3.2. Stratification Analysis
Subsequently, we explored the association between rs8173 G>C polymorphism and WT risk by stratified analysis. The results of the stratified analysis were based on the age, gender, and clinical stage (Table 2). We found that carriers of rs8173 GC/CC genotypes had a decreased WT risk when compared with GG genotype carriers, among children older than 18 months (AOR = 0.56, 95% CI = 0.34–0.93, P=0.024), male children (AOR = 0.54, 95% CI = 0.33–0.90, P=0.017), and those with the clinical stage III + IV diseases (AOR = 0.56, 95% CI = 0.35–0.90, P=0.017).
Table 2.
Stratification analysis of AURKA rs8173 genotypes with Wilms tumor susceptibility.
| Variables | rs8173 (cases/controls) | OR (95% CI) | P | AOR (95% CI)a | P a | |
|---|---|---|---|---|---|---|
| GG | GC/CC | |||||
| Age (months) | ||||||
| ≤18 | 34/97 | 31/136 | 0.65 (0.37–1.13) | 0.127 | 0.65 (0.38–1.13) | 0.128 |
| >18 | 37/99 | 41/199 | 0.55 (0.33–0.91) | 0.021 | 0.56 (0.34–0.93) | 0.024 |
| Gender | ||||||
| Female | 31/92 | 33/141 | 0.70 (0.40–1.21) | 0.199 | 0.70 (0.40–1.22) | 0.203 |
| Male | 40/104 | 39/194 | 0.52 (0.32–0.86) | 0.011 | 0.54 (0.33–0.90) | 0.017 |
| Clinical stages | ||||||
| I + II | 25/196 | 28/335 | 0.66 (0.37–1.16) | 0.144 | 0.71 (0.40–1.26) | 0.245 |
| III + IV | 41/196 | 40/335 | 0.57 (0.36–0.91) | 0.019 | 0.56 (0.35–0.90) | 0.017 |
OR, odds ratio; CI, confidence interval; AOR, adjusted odds ratio. aAdjusted for age and gender, without the corresponding stratification factor.
3.3. Haplotype Analysis
Based on a combined analysis of the three SNP polymorphisms in the AURKA gene, eight haplotypes were inferred (Table 3). Compared with the reference CAC haplotype, only CAG haplotype was associated with significant increased WT risk (AOR = 1.99, 95% CI = 1.05–3.77, P=0.034).
Table 3.
Frequency of inferred haplotypes of the AURKA gene based on observed genotypes and their association with the risk of Wilms tumor.
| Haplotypesa | Cases (n = 286) | Controls (n = 1062) | Crude OR (95% CI) | P | AOR (95% CI)b | P b |
|---|---|---|---|---|---|---|
| CAC | 62 (21.68) | 279 (26.27) | 1.00 | 1.00 | ||
| CAG | 17 (5.94) | 36 (3.39) | 2.00 (1.06–3.77) | 0.033 | 1.99 (1.05–3.77) | 0.034 |
| CTC | 18 (6.29) | 86 (8.10) | 0.89 (0.50–1.57) | 0.676 | 0.91 (0.51–1.62) | 0.748 |
| CTG | 153 (53.50) | 533 (50.19) | 1.21 (0.88–1.68) | 0.240 | 1.21 (0.88–1.67) | 0.249 |
| TAC | 8 (2.80) | 34 (3.20) | 1.00 (0.44–2.25) | 0.990 | 0.99 (0.44–2.24) | 0.983 |
| TAG | 2 (0.70) | 11 (1.04) | 0.77 (0.17–3.55) | 0.736 | 0.74 (0.16–3.40) | 0.694 |
| TTC | 2 (0.70) | 8 (0.75) | 1.06 (0.22–5.09) | 0.945 | 1.09 (0.23–5.28) | 0.913 |
| TTG | 24 (8.39) | 75 (7.06) | 1.35 (0.79–2.30) | 0.266 | 1.34 (0.79–2.29) | 0.279 |
OR, odds ratio; CI, confidence interval; AOR, adjusted odds ratio. aThe haplotype order was rs1047972, rs2273535, and rs8173; bobtained in logistic regression models with adjustment for age and gender.
3.4. Genotype-Based mRNA Expression Analysis
We found that the rs8173 G>C polymorphism was significantly associated with altered AURKA gene expression in transformed fibroblast cells (P=4.3 ∗ 10−12) using data from GTEx portal (Figure 1).
Figure 1.
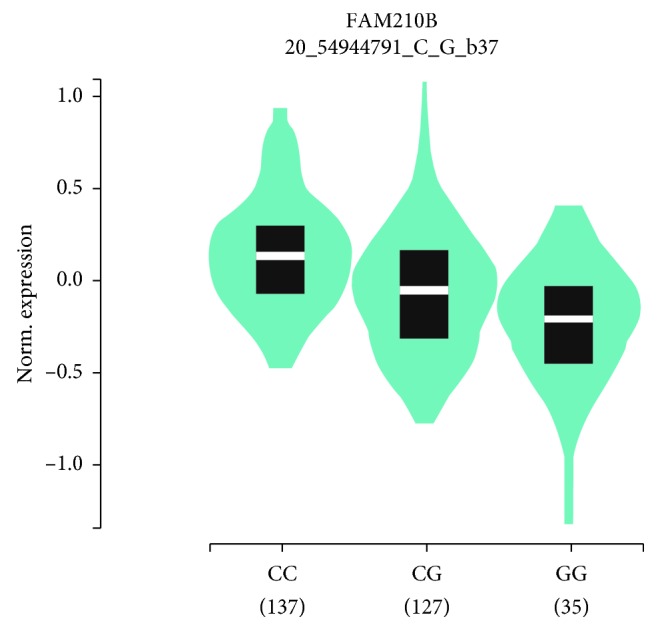
Genotype-based mRNA expression alteration in transformed fibroblasts cells for AURKA rs8173 G>C polymorphism based on data from the GTEx portal database (https://www.gtexportal.org/home/).
4. Discussion
In this case-control study, we analyzed 143 WT patients and 531 controls to investigate the association between three AURKA SNPs (rs1047972, rs2273535, and rs8173) and WT risk of in Chinese children. To the best of our knowledge, this is the first study to investigate the association between AURKA gene polymorphisms and WT susceptibility.
Previous studies have found that AURKA was overexpressed in several common human malignancies, which in turn promoted cell proliferation and tumor progression and metastasis [33–35]. The abnormal expression of AURKA could lead to abnormal chromosome segregation, decreased chromosome stability, and finally increase the susceptibility to malignant transformation [36]. Studies in recent years have shown that AURKA SNPs were closely related to cancer risk. An early study by Lee et al. indicated that the AURKA gene rs2273535 was associated with oral cancer risk [28]. A research by Guo et al. [37] illustrated that the rs2273535 polymorphism was closely related to the increased risk of breast cancer, especially in Asian populations. Interestingly, studies have also found that the Phe allelic variation in AURKA rs2273535 appears to prevent breast cancer in Malaysian Chinese [38]. In addition, Dai et al. [26] reported that the AURKA rs1047972 polymorphism was associated with reduced incidence of breast cancer in Caucasians. These findings suggest that the effects of AURKA SNPs might be tissue dependent and ethnicity dependent.
We previously did not find a significant association between AURKA SNPs and neuroblastoma susceptibility [31]. Interestingly, in this study, our results showed that AURKA rs8173 G>C significantly reduced the WT risk, although no significant association was found for AURKA rs1047972 C>T and rs2273535 T>A. In addition, stratified analysis revealed that individuals carrying AURKA rs8173 GC/CC genotypes had significantly decreased susceptibility to WT in several subgroups, including older than 18 months, male, and clinical stages III + IV.
A previous study showed that the AURKA rs2273535 polymorphism had a strong LD with the rs1047972 genotype. Patients with AURKA haplotype variants exhibited high kinase activity and tended to develop advanced gastric cancer more readily [39]. In this study, we found that the WT risk in individuals with rs1047972/rs2273535/rs8173 CAG haplotype almost doubled, when compared with individuals with CAC haplotype. However, there were no significant association between the other six haplotypes and the risk of WT.
AURKA has been extensively investigated in neuroblastoma. ShRNA-mediated AURKA gene silencing assays have demonstrated that reducing AURKA expression would inhibit cell proliferation in neuroblastoma [40–42]. It is worth mentioning that studies also have found that overexpression or expansion of AURKA is associated with poor prognosis in a variety of cancer patients and inhibition of AURKA expression can trigger tumor cell death [21–23]. Previous studies have shown that the LIN28B-RAN-AURKA axis was involved in the development of neuroblastoma, and AURKA, as a confluence of LIN28B-RAN signaling, could further promote cell cycle progression by phosphorylating many cell cycle regulators and stabilizing N-myc protein (encoded by MYCN gene) [41, 42]. The LIN28B-RAN-AURKA-MYCN signaling cascade in the development of neuroblastoma provides a new insight into the molecular mechanism by which AURKA rs8173 reduced the risk of WT. The role of AURKA in the development of Wilms tumor is currently lacking. However, based on literature search, we found that AURKA is involved in a variety of tumors. Because of the universal contributions of AURKA in tumors, we hypothesized that AURKA might be also implicated in WT and therefore performed the current study. Based on previous reports, we speculated that the AURKA rs8173 might reduce the risk of WT by inhibiting the proliferation and migration of tumor cells through the LIN28B-RAN-AURKA-MYCN signaling pathway.
Some shortcomings should be mentioned in our research. First, the study included 145 patients and 531 controls. Because of the limited sample size, some important results may be accidental. Studies with large samples are indispensible to verify our results. Second, only three AURKA SNPs were investigated in the study. Other potentially functional AURKA gene polymorphisms should be studied in the future. Third, the genotype distribution in this hospital-based study might not represent genotype distribution across the general population, which may bias the case-control study to some extent. Finally, functional experiments should be conducted to strengthen the findings in the current study.
5. Conclusions
Overall, our study confirmed that the AURKA rs8173 G>C polymorphism is associated with reduced WT risk in Chinese children. In the future, multicenter collaboration is needed to further expand the sample size from different regions and different races to clarify the impact of AURKA SNPs on the risk of WT more accurately.
Acknowledgments
This study was funded by grants from the Pearl River S&T Nova Program of Guangzhou (no. 201710010086), the National Natural Science Foundation of China (nos. 81803320 and 81560262), the Science and Technology Project of Guangzhou (no. 201804010037), the Guangdong Provincial Key Laboratory of Research in Structural Birth Defect Disease (no. 2019B030301004), and the Basic Applied Study Planning Projects of Yunnan Province (no. 2018FB130).
Abbreviations
- WT:
Wilms tumor
- WT1:
Wilms' tumor protein 1
- TP53:
Tumor protein 53
- CTNNB1:
Catenin beta 1
- AURKA:
Aurora kinase A
- SNP:
Single nucleotide polymorphism
- HWE:
Hardy–Weinberg equilibrium
- OR:
Odds ratio
- CI:
Confidence interval.
Contributor Information
Tiesong Zhang, Email: zts68420@sina.com.
Jing He, Email: hejing198374@gmail.com.
Data Availability
All the data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Authors' Contributions
Tongyi Lu, Li Li, and Jinhong Zhu contributed equally to this work. All authors contributed significantly to this work. TL, JL, AL, WF, and GL performed the research study and collected the samples and data. JH analyzed the data. HX, JH, and TZ designed the research study. TL, LL, and JZ wrote the paper. JH prepared all the tables and figures. In addition, all authors read, reviewed, and approved the final manuscript to be published.
Supplementary Materials
Supplemental Table 1: frequency distribution of selected variables for Wilms tumor cases and cancer-free controls. Supplemental Table 2: polymorphisms captured by the three included AURKA polymorphisms as predicted by SNPinfo online software. Supplemental Figure 1: linkage disequilibrium (LD) analysis for the three included polymorphisms in Han Chinese population consisting of CHB (Han Chinese in Beijing, China) and CHS (Southern Han Chinese) subjects. Supplemental Figure 2: diagram showing all the genotyped samples for the three included polymorphisms.
References
- 1.Hohenstein P., Hastie N. D. LINking microRNAs, kidney development, and Wilms tumors. Genes & Development. 2014;28(9):923–925. doi: 10.1101/gad.242735.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davidoff A. M. Wilmsʼ tumor. Current Opinion in Pediatrics. 2009;21(3):357–364. doi: 10.1097/mop.0b013e32832b323a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cone E. B., Dalton S. S., Van Noord M., Tracy E. T., Rice H. E., Routh J. C. Biomarkers for Wilms tumor: a systematic review. Journal of Urology. 2016;196(5):1530–1535. doi: 10.1016/j.juro.2016.05.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bao P. P., Li K., Wu C. X., et al. Recent incidences and trends of childhood malignant solid tumors in Shanghai, 2002–2010. Zhonghua Er Ke Za Zhi. 2013;51(4):288–294. [PubMed] [Google Scholar]
- 5.Brok J., Treger T. D., Gooskens S. L., van den Heuvel-Eibrink M. M., Pritchard-Jones K. Biology and treatment of renal tumours in childhood. European Journal of Cancer. 2016;68:179–195. doi: 10.1016/j.ejca.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Dome J. S., Graf N., Geller J. I., et al. Advances in Wilms tumor treatment and biology: progress through international collaboration. Journal of Clinical Oncology. 2015;33(27):2999–3007. doi: 10.1200/jco.2015.62.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee S. H., Paik K. H., Sung K. W., et al. Renal function after tandem high-dose chemotherapy and autologous stem cell transplantation in children with Wilms tumor. Pediatric Transplantation. 2011;15(8):855–860. doi: 10.1111/j.1399-3046.2011.01594.x. [DOI] [PubMed] [Google Scholar]
- 8.Sonn G., Shortliffe L. M. Management of Wilms tumor: current standard of care. Nature Clinical Practice Urology. 2008;5(10):551–560. doi: 10.1038/ncpuro1218. [DOI] [PubMed] [Google Scholar]
- 9.Pritchard-Jones K., Moroz V., Vujanić G., et al. Treatment and outcome of Wilms’ tumour patients: an analysis of all cases registered in the UKW3 trial. Annals of Oncology. 2012;23(9):2457–2463. doi: 10.1093/annonc/mds025. [DOI] [PubMed] [Google Scholar]
- 10.Malogolowkin M., Cotton C. A., Green D. M., et al. Treatment of Wilms tumor relapsing after initial treatment with vincristine, actinomycin D, and doxorubicin: a report from the National Wilms Tumor Study Group. Pediatric Blood & Cancer. 2008;50(2):236–241. doi: 10.1002/pbc.21267. [DOI] [PubMed] [Google Scholar]
- 11.Little S. E., Hanks S. P., King-Underwood L., et al. Frequency and heritability of WT1 mutations in nonsyndromic Wilms’ tumor patients: a UK children’s cancer study group study. Journal of Clinical Oncology. 2004;22(20):4140–4146. doi: 10.1200/jco.2004.02.136. [DOI] [PubMed] [Google Scholar]
- 12.Scott R. H., Douglas J., Baskcomb L., et al. Constitutional 11p15 abnormalities, including heritable imprinting center mutations, cause nonsyndromic Wilms tumor. Nature Genetics. 2008;40(11):1329–1334. doi: 10.1038/ng.243. [DOI] [PubMed] [Google Scholar]
- 13.Andrade R. C., Cardoso L. C. A., Ferman S. E., et al. Association of TP53 polymorphisms on the risk of Wilms tumor. Pediatric Blood & Cancer. 2014;61(3):436–441. doi: 10.1002/pbc.24775. [DOI] [PubMed] [Google Scholar]
- 14.Gadd S., Huff V., Walz A. L., et al. A children’s oncology group and TARGET initiative exploring the genetic landscape of Wilms tumor. Nature Genetics. 2017;49(10):1487–1494. doi: 10.1038/ng.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou X., Wang P., Zhao H. The association between AURKA gene rs2273535 polymorphism and gastric cancer risk in a Chinese population. Frontiers in Physiology. 2018;9:p. 1124. doi: 10.3389/fphys.2018.01124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan M., Wang C., He B., et al. Aurora-A kinase: a potent oncogene and target for cancer therapy. Medicinal Research Reviews. 2016;36(6):1036–1079. doi: 10.1002/med.21399. [DOI] [PubMed] [Google Scholar]
- 17.Bischoff J. R., Anderson L., Zhu Y., et al. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. The EMBO Journal. 1998;17(11):3052–3065. doi: 10.1093/emboj/17.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tong T., Zhong Y., Kong J., et al. Overexpression of Aurora-A contributes to malignant development of human esophageal squamous cell carcinoma. Clinical Cancer Research. 2004;10(21):7304–7310. doi: 10.1158/1078-0432.ccr-04-0806. [DOI] [PubMed] [Google Scholar]
- 19.Goos J. A. C. M., Coupe V. M. H., Diosdado B., et al. Aurora kinase A (AURKA) expression in colorectal cancer liver metastasis is associated with poor prognosis. British Journal of Cancer. 2013;109(9):2445–2452. doi: 10.1038/bjc.2013.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ewart-Toland A., Briassouli P., de Koning J. P., et al. Identification of Stk6/STK15 as a candidate low-penetrance tumor-susceptibility gene in mouse and human. Nature Genetics. 2003;34(4):403–412. doi: 10.1038/ng1220. [DOI] [PubMed] [Google Scholar]
- 21.Marumoto T., Honda S., Hara T., et al. Aurora-A kinase maintains the fidelity of early and late mitotic events in HeLa cells. Journal of Biological Chemistry. 2003;278(51):51786–51795. doi: 10.1074/jbc.m306275200. [DOI] [PubMed] [Google Scholar]
- 22.Gorgun G., Calabrese E., Hideshima T., et al. A novel Aurora-A kinase inhibitor MLN8237 induces cytotoxicity and cell-cycle arrest in multiple myeloma. Blood. 2010;115(25):5202–5213. doi: 10.1182/blood-2009-12-259523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou N., Singh K., Mir M. C., et al. The investigational Aurora kinase A inhibitor MLN8237 induces defects in cell viability and cell-cycle progression in malignant bladder cancer cells in vitro and in vivo. Clinical Cancer Research. 2013;19(7):1717–1728. doi: 10.1158/1078-0432.ccr-12-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niu H., Shin H., Gao F., et al. Aurora a functional single nucleotide polymorphism (SNP) correlates with clinical outcome in patients with advanced solid tumors treated with alisertib, an investigational aurora a kinase inhibitor. EBioMedicine. 2017;25:50–57. doi: 10.1016/j.ebiom.2017.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu L., Zhou X., Jiang F., Xu L., Yin R. STK15 rs2273535 polymorphism and cancer risk: a meta-analysis of 74,896 subjects. Cancer Epidemiology. 2014;38(2):111–117. doi: 10.1016/j.canep.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 26.Dai Z. J., Kang H. F., Wang X. J., et al. Association between genetic polymorphisms in AURKA (rs2273535 and rs1047972) and breast cancer risk: a meta-analysis involving 37,221 subjects. Cancer Cell International. 2014;14(1):p. 91. doi: 10.1186/s12935-014-0091-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang B., Hsu C.-J., Chou C.-H., et al. Variations in the AURKA gene: biomarkers for the development and progression of hepatocellular carcinoma. International Journal of Medical Sciences. 2018;15(2):170–175. doi: 10.7150/ijms.22513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee C.-P., Chiang S.-L., Lee C.-H., et al. AURKA Phe31Ile polymorphism interacted with use of alcohol, betel quid, and cigarettes at multiplicative risk of oral cancer occurrence. Clinical Oral Investigations. 2015;19(8):1825–1832. doi: 10.1007/s00784-015-1432-5. [DOI] [PubMed] [Google Scholar]
- 29.Fu W., Zhu J., Xiong S.-W., et al. BARD1 gene polymorphisms confer nephroblastoma susceptibility. EBioMedicine. 2017;16:101–105. doi: 10.1016/j.ebiom.2017.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He J., Qiu L.-X., Wang M.-Y., et al. Polymorphisms in the XPG gene and risk of gastric cancer in Chinese populations. Human Genetics. 2012;131(7):1235–1244. doi: 10.1007/s00439-012-1152-8. [DOI] [PubMed] [Google Scholar]
- 31.Tang J., Qian Y., Zhu J., et al. Lack of associations between AURKA gene polymorphisms and neuroblastoma susceptibility in Chinese children. Bioscience Reports. 2018;38(3) doi: 10.1042/bsr20180292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu J., Fu W., Jia W., Xia H., Liu G.-C., He J. Association between NER pathway gene polymorphisms and Wilms tumor risk. Molecular Therapy—Nucleic Acids. 2018;12:854–860. doi: 10.1016/j.omtn.2018.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Treekitkarnmongkol W., Katayama H., Kai K., et al. Aurora kinase-A overexpression in mouse mammary epithelium induces mammary adenocarcinomas harboring genetic alterations shared with human breast cancer. Carcinogenesis. 2016;37(12):1180–1189. doi: 10.1093/carcin/bgw097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou H., Kuang J., Zhong L., et al. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nature Genetics. 1998;20(2):189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 35.Xu L. Z., Long Z. J., Peng F., et al. Aurora kinase a suppresses metabolic stress-induced autophagic cell death by activating mTOR signaling in breast cancer cells. Oncotarget. 2014;5(17):7498–7511. doi: 10.18632/oncotarget.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sen S., Zhou H., White R. A. A putative serine/threonine kinase encoding gene BTAK on chromosome 20q13 is amplified and overexpressed in human breast cancer cell lines. Oncogene. 1997;14(18):2195–2200. doi: 10.1038/sj.onc.1201065. [DOI] [PubMed] [Google Scholar]
- 37.Guo X.-G., Zheng L., Feng W.-B., Xia Y. The AURKA gene rs2273535 polymorphism contributes to breast carcinoma risk—meta-analysis of eleven studies. Asian Pacific Journal of Cancer Prevention. 2014;15(16):6709–6714. doi: 10.7314/apjcp.2014.15.16.6709. [DOI] [PubMed] [Google Scholar]
- 38.Chong E. T. J., Goh L. P. W., See E. U. H., Chuah J. A., Chua K. H., Lee P.-C. Association of CYP2E1, STK15 and XRCC1 polymorphisms with risk of breast cancer in Malaysian women. Asian Pacific Journal of Cancer Prevention. 2016;17(2):647–653. doi: 10.7314/apjcp.2016.17.2.647. [DOI] [PubMed] [Google Scholar]
- 39.Ju H., Cho H., Kim Y. S., et al. Functional polymorphism 57Val>Ile of Aurora kinase A associated with increased risk of gastric cancer progression. Cancer Letters. 2006;242(2):273–279. doi: 10.1016/j.canlet.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 40.Cole K. A., Huggins J., Laquaglia M., et al. RNAi screen of the protein kinome identifies checkpoint kinase 1 (CHK1) as a therapeutic target in neuroblastoma. Proceedings of the National Academy of Sciencesof the United States of America. 2011;108(8):3336–3341. doi: 10.1073/pnas.1012351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schnepp R. W., Khurana P., Attiyeh E. F., et al. A LIN28B-RAN-AURKA signaling network promotes neuroblastoma tumorigenesis. Cancer Cell. 2015;28(5):599–609. doi: 10.1016/j.ccell.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Otto T., Horn S., Brockmann M., et al. Stabilization of N-Myc is a critical function of Aurora A in human neuroblastoma. Cancer Cell. 2009;15(1):67–78. doi: 10.1016/j.ccr.2008.12.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1: frequency distribution of selected variables for Wilms tumor cases and cancer-free controls. Supplemental Table 2: polymorphisms captured by the three included AURKA polymorphisms as predicted by SNPinfo online software. Supplemental Figure 1: linkage disequilibrium (LD) analysis for the three included polymorphisms in Han Chinese population consisting of CHB (Han Chinese in Beijing, China) and CHS (Southern Han Chinese) subjects. Supplemental Figure 2: diagram showing all the genotyped samples for the three included polymorphisms.
Data Availability Statement
All the data used to support the findings of this study are available from the corresponding author upon request.


