Abstract
The neuronal growth cone is a highly motile, specialized structure for extending neuronal processes. This structure is essential for nerve growth, axon pathfinding, and accurate synaptogenesis. Growth cones are important not only during development but also for plasticity-dependent synaptogenesis and neuronal circuit rearrangement following neural injury in the mature brain. However, the molecular details of mammalian growth cone function are poorly understood. This review examines molecular findings on the function of the growth cone as a result of the introduction of novel methods such superresolution microscopy and (phospho)proteomics. These results increase the scope of our understating of the molecular mechanisms of growth cone behavior in the mammalian brain.
Keywords: axon guidance, proteomics, phosphorylation, superresolution microscopy, lipid rafts, chondroitin sulfate
1. Introduction
The neuronal growth cone is a highly motile structure that is specifically formed at the tips of developing axons and dendrites. This structure was discovered and first described by the distinguished anatomist Santiago Ramón y Cajal, who was the first neuroscientist, along with Camillo Golgi, to win a Nobel prize.1) This review mainly focuses on the axonal growth cone, because the dendritic growth cone is not well characterized.
A pioneering study using cell culture revealed that the growth cone actually moves when the neurite is growing,2) and a live-imaging study (movie) by Junnosuke Nakai revealed that growth cone movement is regulated by multiple cues.3) Molecular and cellular biology studies have shown that F-actin initiates growth cone motility,4) and genetic approaches to neural wiring5,6) have contributed to the molecular understanding of growth cone behavior. Axon guidance molecules, such as netrin, semaphorins, and ephrins, are localized outside the growth cone, and signaling in the growth cone occurs in response to such molecules. In addition, each guidance molecule frequently induces opposite movements in the direction of the growth cone, i.e., attraction or repulsion. Thus, determination of growth cone direction is dependent upon the signaling in it rather than the guidance molecules themselves.
The fundamental mechanisms of other neural development processes, such as regional determination, neural cell lineage, neurogenesis, neuronal migration, and synaptogenesis in the mammalian central nervous system (CNS), are relatively well understood due to investigations over the last quarter century. However, the fundamental molecular machinery involved in growth cone functions in the brain of higher organisms remains poorly understood.
Most growth cone studies have been primarily performed using (1) model organisms, such as Caenorhabditis elegans or Drosophila; (2) other invertebrate neurons, such as those from Aplysia; or (3) peripheral nerves of vertebrates, such as chick dorsal root ganglion (DRG) neurons. These studies have contributed to progress in this field when molecular and cellular neurobiological techniques using mammalian CNS neurons are not sufficiently feasible. However, the results derived from model organisms or the chick DRG cannot be simply applied to the understanding of molecular pathways in the mammalian CNS growth cone, because mammalian signaling mechanisms are generally much more complex and have greater diversity than those of simpler neuronal systems. To approach this problem, we utilized methods that are readily available for directly determining the numerous protein components in the growth cone of the mammalian CNS.
Currently, mammalian CNS growth cones are the most intensively studied. Clearly, the growth cones of simpler organisms or those outside the CNS (such as DRG) have completely different molecular compositions and interactions compared with the mammalian CNS. Simple hypotheses have been proposed from studies using non-mammalian growth cones. These hypotheses, especially the molecular aspects, should be tested to determine whether they apply to the mammalian CNS. Over the last decade, mammalian CNS neurons, such as cortical or hippocampal neurons, have been utilized to discover additional molecular signaling pathways. Because hippocampal and DRG neurons differ considerably from each other,7) the molecular mechanisms of their growth cone behavior are also likely to be considerably different.
Recently, new techniques have been introduced in studies on the molecular mechanisms of mammalian growth cone behavior, and this review summarizes the results of the new findings.
2. Basic structure and signaling of the growth cone
In the growth cone, microtubules (MTs) and various vesicles are concentrated near distal axonal shafts, a region called the central (C-) domain of the growth cone.8) These vesicles function as plasmalemmal precursors for axonal growth, although the details of their turnover and recycling are not well understood.9) In addition, some vesicles may be endosomes derived from endocytosis.10) Although morphological vesicular categorization in growth cones was reported previously using electron microscopy, molecular markers that discriminate among these have not yet been determined.8,11,12) Vesicular trafficking in growth cones is important not only for maintenance of the plasma membrane but also for axonal guidance.11,13–15)
The area near the leading edge of the growth cone, called the peripheral (P-) domain,8) has a high concentration of dynamic F-actin, and only this domain enables the growth cone to move. The coordination between F-actin rearrangement in the P-domain and MT growth in the C-domain is essential for growth cone function, but the molecules that link these two have not been defined exactly.16,17) In the P-domain, branched F-actin forms lamellipodia, whereas unbranched filaments form slender protrusions, called filopodia. In Aplysia growth cones, which are much larger than mammalian ones, a transition zone (T-zone) is clearly discriminated between the C- and P-domains.18,19) However, this zone is not distinctly recognized in mammalian growth cones. This review will not discuss the T-zone further.
In the C-domain, the cortical membrane skeleton, which is composed of F-actin, spectrin, and other components (discussed below13)), is enriched underneath the plasma membrane. These structures are likely important for changes in the structure of the growth cone when the axon grows and the new MTs invade the growth cone area. However, F-actin dynamics and the molecular mechanisms involved in this process have not yet been analyzed.
The distribution of organelles in the growth cone has not been described clearly, even using electron microscopy. Growth cones contain mitochondria, a few clusters of ribosomes, branched membranous reticulum, lysosomes, cytoskeleton,16) membranous disks, and vacuoles.17) Some of these structures have been demonstrated by identification of molecular markers using proteomics.13,20) Local protein synthesis by the ribosomes in growth cones has been demonstrated and may be involved in determination of their advancing direction21) (see below section 3.4).
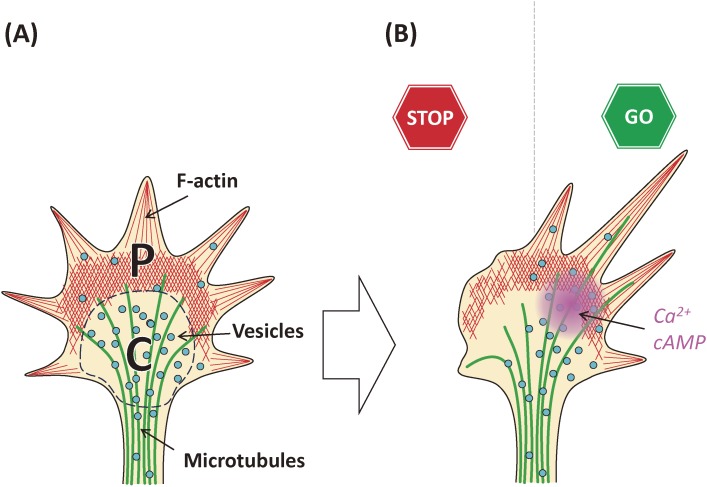
Two second messengers are present in the growth cone, Ca2+ and cAMP12,22–24) (see also a review;25) Fig. 1). These second messengers are involved in signaling induced by axon guidance molecules such as netrin, semaphorins, or ephrins.12,25) Diffusible factors are also attached to the cell surface/extracellular matrix to guide the growth cone. Thus, in vivo signaling including local translation mechanisms should be revisited, and the classical gradient theory should be revised.26,27)
Figure 1.
The growth cone structure (A) and its behaviors are regulated by second messengers (B). The growth cone is divided into two regions; one is the central (C-) domain in which MTs and vesicles are enriched; the other is the peripheral (P-) domain in which dynamic F-actin bundles are enriched, forming lamellipodia and filopodia. Growth cone behavior is regulated by axon guidance molecules via up- and down-regulation of second messengers such as Ca2+ and cAMP. Axon guidance molecules are either attractive (“GO”) or repulsive (“STOP”) to the growth cone, probably via higher and lower concentrations of second messengers for attractive and repulsive directions, respectively.
3. Proteomics profiling of the mammalian growth cone
As mentioned above (see 1. Introduction), complete molecular profiling of the mammalian CNS growth cone is needed for understanding of the molecular basis of growth cone function. Proteomics analysis, which quantitatively identifies numerous proteins, has completely overcome previous methodological limitations for determining protein components.28) We performed a proteomics study of mammalian growth cones collected by subcellular fractionation.29–32) Following subcellular fractionation, isolated growth cones or growth cone particles (GCPs) were enriched, and hypotonic treatment of this fraction provides growth cone membranes (GCMs).31) Using these methods, we characterized many previously unknown growth cone proteins.20) After our report, another group identified additional GCP proteins by proteomics,33) and almost all of our results were validated in their report. Surprisingly, although we performed immunostaining for more than 200 proteins, no false positive cases were found. These results confirmed the extremely high purity of our GCP/GCM fractions.20)
In contrast to synapse-associated proteins in mature synapses,34–36) molecular markers for growth cones, except for the classical neuronal growth-associated protein, GAP-43, have not been established.37) We identified approximately 20 proteins that are functional molecular markers of the growth cone; these were termed neuronal growth-associated proteins (nGAPs) and are involved in the functions of mammalian CNS growth cones.20) Initially, we were slightly surprised that few proteins were found compared with the number involved in neurite growth in C. elegans or Drosophila. The reason for this difference was that these proteins in lower organisms are not generally concentrated in their nervous systems in contrast to the mammalian CNS. This finding suggested that the molecular mechanisms of mammalian neuronal growth differ considerably from those of simple organisms.
Because all putative nGAPs from cortical neurons satisfied the criteria of “nGAPs”, even in PC12D cells,38) nGAPs (Fig. 2) are considered universal growth cone markers that are not dependent on neuronal species. Recently, some proteomics studies of developing neurons have been published.39,40) They included information about locally synthesized proteins. These data will be useful for understanding the molecular aspects of growth cone functions. In addition, the proteomics techniques employed by another group also were also useful for identification of a specific new protein called shootin-1, which regulates F-actin during axon growth.41) Characteristics of the growth cone proteins identified by proteomics have been described in detail previously.13) Thus, only recently identified proteins are summarized here.
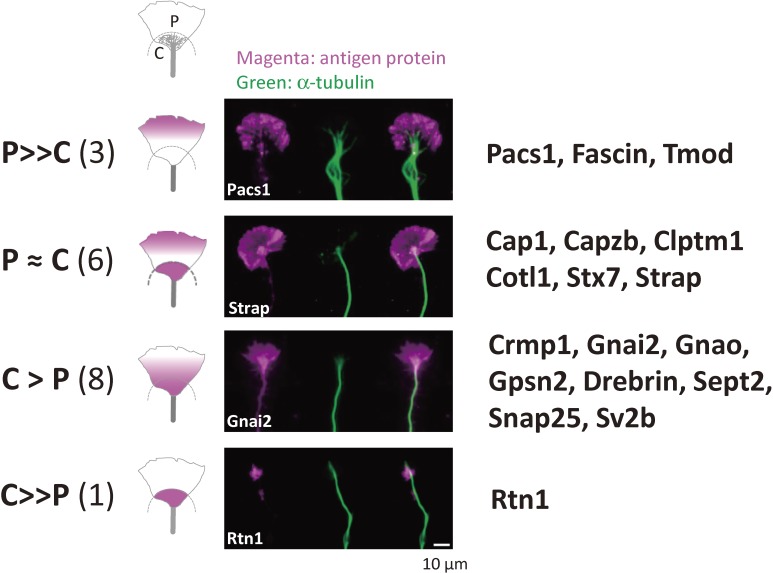
Figure 2.
The nGAPs identified in our proteomics studies.20) C: central domain; P: peripheral domain. P ≫ C, P-domain-dominant localization; P ≈ C; evenly distributed both in C- and P-domains; C > P, enriched in the C-domain compared to the P-domain; C ≫ P, C-domain-dominant localization.
3.1. MTs and MT-related proteins.
Two types of MT-binding proteins are present in the growth cone. Microtubule-associated proteins (MAPs) are localized adjacent to MTs and stabilize them. MAP1B and tau are mainly localized in the mammalian growth cone.20) MAP6, also called STOP protein, was identified as a MAP with growth cone proteomics,20) and this protein is involved in rapid axon growth for neuronal polarity determination through palmitoylation42) (discussed below). However, curiously, this protein has functions that are independent of MT binding. Through its binding to SH3 domain-containing proteins, MAP6 also acts as a downstream signaling protein of semaphorin-3E.43)
Plus-end interacting proteins (+TIPs) of MTs are a completely different protein group that selectively bind to MT (+)-ends at selected polymerization sites distinct from MAP-binding sites. +TIPs are composed of many subgroups, each of which is structurally different.44) These proteins are considered strong candidates for proteins that link MTs and F-actin in the growth cone.45) TACC3 is a minor +TIP that promotes axon elongation.46)
3.2. Actin filaments and actin-binding proteins.
As described previously, filopodia and lamellipodia, in which the dynamic actin filaments are concentrated in the P-domain, are present in the growth cone. The actin polymerization and depolymerization cycle is believed to determine the direction of axon growth.47) By proteomics analysis of the growth cone, we characterized the actin-binding proteins.13,20) The classical idea of growth cone motility based upon filopodial F-actin is summarized in several reviews.48) Three types of actin isoforms, α-, β-, and γ-actin, are localized in the growth cone.20) α- and β-actin are involved in axonal filopodia dynamics and collateral branch formation, and dynamic movements of growth cone filopodia, respectively.49) The shapes of the growth cone are generally regulated by F-actin organization.50–53)
The single-headed myosin, Myo1b, which is present in the growth cone,13,20) is a necessary component for axon growth via actin waves from the cell body.54) Reports disagree regarding whether actin waves are physiologically involved in neuronal development or not;55,56) thus, further studies are needed to investigate this idea.
3.3. Signaling proteins.
3.3.1. Protein kinases.
Our proteomics data15) showed that protein kinase A (PKA) and Ca2+/calmodulin-dependent protein IIβ (CaMKIIβ) are the major protein kinases in the growth cone. Thus, as mentioned above, cAMP and Ca2+ are the postulated second messengers in the growth cone. However, how these second messengers are increased and what types of biochemical mechanisms are involved are not yet clear. For example, the levels of voltage-gated Ca2+ channels are much lower than in mature synapses, and most Ca2+ channels in the growth cone are L-type channels.20) A recent report using a new Ca2+ indicator supported the idea that this type of Ca2+ channel is important for nerve growth.57)
3.3.2. Heterotrimeric G proteins (G proteins) and G-protein coupled receptors (GPCRs).
Heterotrimeric G-proteins are highly concentrated in the growth cone.13,20,58–60) However, the significance of G-proteins has not been characterized well due to the low abundance of GPCRs.13,61) The human autosomal recessive neurological disorder Chudley–McCullough syndrome is caused by defective Gpsm2/Gαi3 signaling in stereocilia, which causes disruption of actin dynamics in the growth cone.62) The precise signaling pathways upstream and downstream of GPCRs should be analyzed regarding growth cone functions. In addition, the GPCR latrophilin-1 is involved in short-range axon pathfinding by interacting with the proteolytic fragment of the membrane protein Lasso/tenurin-2.63) Latrophilin-1 is a presynaptic adhesion GPCR protein in the mature neuron,64) but this protein is also present in the growth cone.13,20) How the G-proteins are related to this process may be important for local axon attraction/repulsion.
3.3.3. Rab family.
Many Rab proteins are present in growth cones,20) and some reports suggest that they are involved in nerve growth.65,66) However, their direct functions or interactions regarding axon growth have not been clarified.67) In contrast to other small GTPases (Ras/Rho family proteins) in the growth cone, the activation and inactivation mechanisms of these Rabs, such as their activators (guanine nucleotide exchange factors) or inactivators (GTPase-activating proteins), are not known. Rab11, a recycling marker, supports axon regeneration;68) thus, how Rab proteins work in the growth cone should be studied further.
3.3.4. Adapter molecules.
(a) CRMP family. CRMP2 was first characterized as a neuronal polarity-dependent protein,69) and CRMP4 is involved in cytoskeletal rearrangement for axon growth/regeneration.70,71) The CRMP family includes the most important and abundant adapter molecules. In addition, CRMP2 interacts with Sra-1/CYFIP, which is a Rac-associated regulator of the cytoskeleton, and regulates axon formation.72) Because CYFIP is an nGAP,20) this report is quite interesting.
(b) 14-3-3 family members. The 14-3-3 family includes adapter proteins for proteins phosphorylated by serine (S)/threonine (T) kinases, in particular, PKA.73) Among the many members of this family, we confirmed that β, ε, ζ, η, γ, and θ forms are abundantly present in GCPs. 14-3-3-dependent signaling is thought to be a major event downstream of PKA in the growth cone,74) and a chemical compound fusicoccin-A is a stimulator of axon regeneration.75)
3.3.5. Protein translation machinery (Fig. 3).
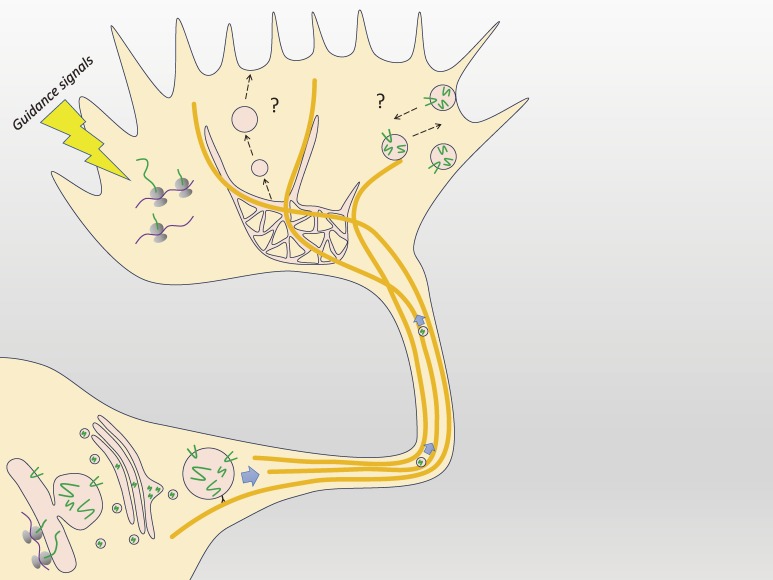
Figure 3.
Protein usage in the growth cone. Proteins used in the growth cone for axon growth are mainly transported axonally after their synthesis in the cell body. However, some proteins are locally synthesized in the growth cone in response to axon guidance signals. Transported vesicular proteins probably undergo recycling, although most of the precise mechanisms and dynamics of these processes are uncharacterized.
The growth cone has the potential for local protein synthesis, including initiation factors, elongation factors, and ribosomal components.13,20) Local protein synthesis there is believed to modify local axon guidance,12) but not axonal growth itself.76) This hypothesis is supported by the observation of anterograde axonal transport derived from massive protein synthesis in the cell body. These large amounts of proteins are probably directly utilized for axon growth. In Xenopus, many locally synthesized proteins in developing axons have been identified.76)
3.3.6. Protein degradation systems.
Most TRIM family members are known as E3 ubiquitin ligases. TRIM9 is an E3 ligase that is involved in filopodial actin reorganization.77) TRIM46 lacks ubiquitin ligase activity, but this protein interacts with MTs and controls MT reorganization.78)
3.3.7. Proteins involved in exocytosis.
Our group and others have shown that nerve growth is mediated by the SNARE proteins and related proteins.79–82) These proteins are most likely involved in vesicular fusion and targeting in the growth cone.9)
Syntaxin-1 is involved in axon guidance through guidance molecules83–86) and is conserved in a wide range of organisms.87) Many types of syntaxin-binding proteins have been characterized,35) including its most abundant partner, Munc-18-1, in the growth cone.20) Other partners for axon growth are not well characterized.
4. Phosphoproteomics reveals new signaling pathways in the growth cone
4.1. General profiling of GCM phosphoproteomics.
To further investigate molecular signaling in growth cones, this review will now focus on protein phosphorylation, the most important regulatory mechanism in many cellular processes88) and in physiological and pathological conditions in neuroscience.89) Many reports about phosphorylation-dependent pathways in the growth cone have been published.90) However, most studies have used in vitro phosphorylation systems that do not necessarily represent the in vivo situation. In addition, ∼500 protein kinases are encoded in mammalian genomes, and determination of the specific activating kinase is difficult.91) Currently, phosphoproteomics is an important and powerful method for comprehensive and quantitative identification of in vivo phosphorylation sites.92–94)
We performed phosphoproteomics analyses of GCMs31) and identified more than 30,000 phosphopeptides and ∼4,600 different phosphorylation sites from ∼1,200 proteins.95) These phosphorylation sites are likely those that are necessary for in vivo growth cone activity. Surprisingly, most of the highly frequent phosphorylation sites (>100 times) were previously uncharacterized.95) These results indicate that a large number of unknown, potentially important phosphorylated sites are involved in mammalian growth cone functions.
S or T protein kinases are categorized into four classes: acidic, basic, proline (P)-directed, and other kinases, based on the neighboring amino acid residues of the phosphorylated S or T.96) We showed that more than 60% of S or T phosphorylation sites in GCMs were P-directed phosphorylation. We applied bioinformatics analyses to these data and revealed that these frequently activated P-directed kinases were mitogen-activated protein kinases (MAPKs).95) The MAPK family is composed of three subfamilies: extracellular signal-regulated kinase, c-Jun N-terminal kinase (JNK), and p38. Inhibitor studies determined that JNK is the most likely kinase responsible for the highly frequent phosphorylation sites.95) This was unexpected because JNK is an apoptotic signal in many cells,97) and in neurons, JNK induces axon degeneration.98)
We have reported that at least seven highly phosphorylated sites in MAP1B (two sites), GAP-43 (two sites), SCG10,99) Rufy3,100,101) and Robo2 are JNK dependent.95) These results indicated that JNK activation is physiologically required in the mammalian growth cone via phosphorylation of proteins that belong to the “actual working unit” for axon growth (Fig. 4).
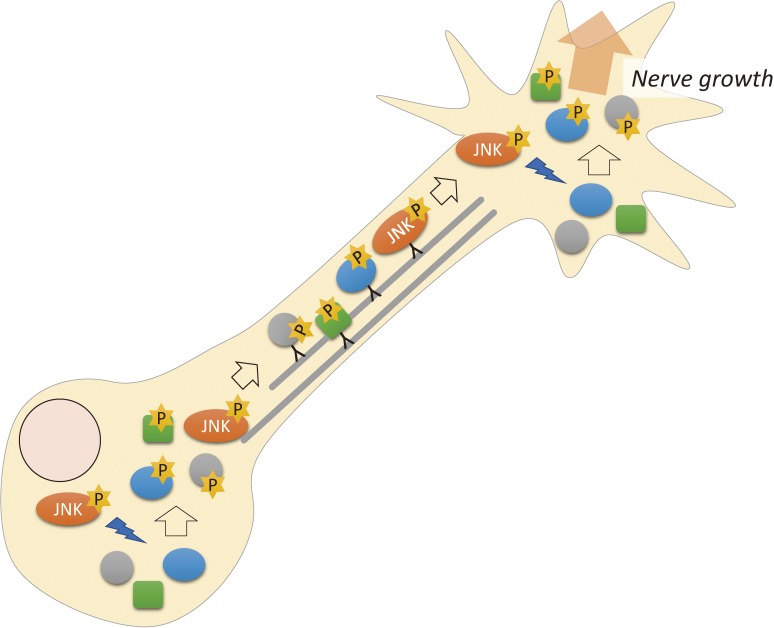
Figure 4.
JNK probably phosphorylates various proteins that are directly necessary for neuronal growth. JNK is activated in the cell bodies, but active JNK may undergo anterograde axonal transport. Phosphorylated proteins are also thought to be transported from the cell body to the growth cone, although some may be phosphorylated locally in the growth cone.
JNK phosphorylates many kinds of proteins that are directly involved in axon growth/regeneration.95) A decade ago, reports described that normal brain development requires the activation of JNK,102–106) and adult neuronal plasticity also involves JNK activity.107–109) JNK plays a role in retrograde axonal transport and phosphorylates transcription factors in the nucleus.110) However, our results on phosphoproteomics revealed that JNK phosphorylates many proteins involved in axon formation.
Several lines of evidence suggest that the developing neuron has high JNK activity, which is essential for the physiological development of the mammalian brain.106,111) In addition, axon guidance and regeneration, which are dependent on netrin-1, likely also require JNK activation.112,113) Why the high enzymatic activity of JNK does not induce apoptosis is not clear. Our phosphoproteomics results provided an unbiased demonstration that nerve growth during development in the mammalian brain requires JNK activity and involves a large number of its substrates, which belong to various categories.
4.2. Phosphorylation of GAP-43.
In our GCM phosphoproteomics study, the most abundant phosphorylated site was S96 of the classical growth cone marker GAP-43,95,114,115) comprising more than 1% of all phosphopeptides. This phosphorylated site had only been reported once, more than a quarter of a century ago,116) just before the discovery of JNK itself.117) As a result, S96 and JNK were never related to each other until our study.95) S96 phosphorylation (pS96) is JNK dependent. A pS96-Ab specifically recognizes growing axons in vivo during certain developmental stages.95) In addition, pS96 is detected in regenerating axons after sciatic injury, not only by immunohistochemistry,95,118) but also directly by mass spectrometry.95)
GAP-43/neuromodulin was one of the most featured proteins in vertebrate axon growth/regeneration before its knockout (KO) was produced.119) Contrary to the expectation from cellular findings, no large changes in nerve growth or neuronal connections were found in the KO line. The phenotype of these mice reduced researchers’ interest in this protein, and this protein has received little attention using the new technologies in neuroscience.
GAP-43/neuromodulin has several unique biochemical properties. (1) This protein is an a intrinsically disordered protein. The protein does not have specific high-order structures, but acquires specific functions or interactions after modifications such as phosphorylation. A recent nuclear magnetic resonance study120) confirmed this idea for GAP-43. (2) This protein is palmitoylated (C3C4) and attached to membranes, probably to the plasma membrane.121) (3) GAP-43/neuromodulin is the first characterized calmodulin-binding protein, via its IQ domain/motif.115)
Protein kinase C-dependent S41 phosphorylation of GAP-43, a previous area of study, has not been identified in the growth cone.95) This site is not detected in regenerating axons; thus, we concluded that its phosphorylation does not direct axon growth/regeneration, although a number of studies referred to these relationships more than 20 years ago.
Unsolved: Why are a large number of growth cone proteins, which should support nerve growth, simultaneously phosphorylated by JNK? This is the most important problem to solve.
5. Superresolution microscopy reveals associations between F-actin organization and membrane trafficking in growth cones
To better understand the molecular basis of neuronal growth, the precise relationship between both cytoskeletal and membrane components must be clarified. In addition, the mammalian CNS growth cone is three-dimensional, compared with those of the peripheral nervous system or invertebrate neurons, the growth cones of which are flat. However, little progress has been made despite the 3D elucidation of the growth cone structure. Unfortunately, the diffraction limitation of conventional optical microscopy (∼200 nm), including conventional confocal microscopy, precludes such analyses.122,123) Several types of superresolution microscopy (structured illumination microscopy [SIM], stimulated emission depletion microscopy [STED], photoactivation localization microscopy [PALM], stochastic optical reconstruction microscopy [STORM], etc.), which are based upon different principles, have succeeded in overcoming the optical diffraction limit and have achieved a resolution of 50–100 nm.123) Superresolution microscopy has revealed a newly recognized distribution of F-actin in the axon, which is implicated in axonal transport, polymerization of F-actin,124–127) and the distribution of tau and MTs in growth cones.128)
SIM can be used to visualize the fine structure of cells by calculating the interference (moiré) patterns induced by irradiation with striped-pattern excitation light.129) Using SIM, approximately 100 nm in lateral dimensions and 300 nm in the axial dimension can be visualized. Superresolution images are easy to obtain with SIM, because it utilizes typical fluorescent dyes.130,131)
Using SIM, we determined the precise localization of a number of proteins identified by growth cone proteomics.20,132) Our analysis using SIM revealed a new linkage between the organization of F-actin bundles and local endocytosis at the leading edge,132,133) which could never have been acquired using confocal microscopy. Immediately after publication of the proteomics results, we began to analyze vesicular movements for live imaging. However, the dense distributions of growth cone vesicles prevented the measurement of quantitative data using confocal microscopy (Fig. 5).
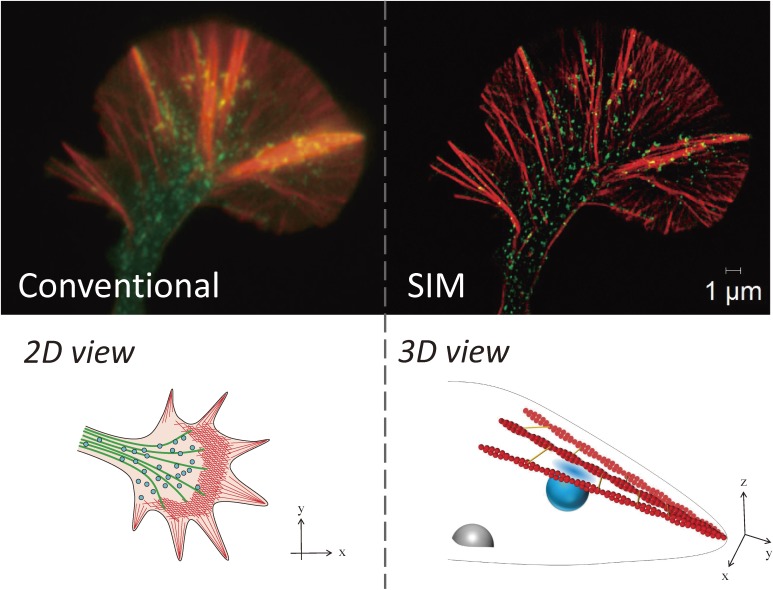
Figure 5.
Superresolution microscopy reveals new endocytotic sites for nerve growth. Red, mCherry-actin; Green, EGFP-synaptophysin. Conventional confocal microscopy view (upper left) is not suitable for visualizing the densely packed cytoskeleton and vesicles. In addition, confocal microscopy is generally used to acquire two-dimensional views (lower left). SIM superresolution can be used for clear, 3D acquisition of the distribution of the growth cone cytoskeleton and vesicles (upper right).
These results have important implications. First, vesicular movements in the P-domain were clarified. From classical studies, vesicles in the C-domain were thought to be fused and undergo recycling.8,15) However, vesicles in the P-domain do not fuse with each other, and they move toward the C-domain.8) These previous studies suggested that vesicles in the C- and P-domains have different dynamics and that their molecular characteristics may be different. Here, we clarified vesicular movement in the P-domain directly and determined the significance. Second, we studied coordinated movements of the vesicles and the actin cytoskeleton for axonal growth in the growth cone. We also clarified the involvement of key molecules in this process, i.e., dynamin, endophilin, and fascin. Fascin is an actin-binding protein that forms F-actin bundles and is one of the nGAPs we identified.20) Third, the newly identified type of endocytosis in the P-domain, which requires dynamin and a BAR domain-containing protein (which regulates membrane curvature), endophilin, is similar to fast endocytosis134) and occurs in the z-axis direction. Finally, some of the F-actin in the P-domain is also distributed three-dimensionally.122,132)
Unsolved: The first problem is where vesicular fusion occurs for plasma membrane expansion in axon growth. If can we observe movement of the growth cone in the z-axis direction, we may find the answer. Second, the significance of recycling for axonal growth is not yet known.
6. Lipid rafts in the growth cone and neuronal polarity signals
In various cells, lipid-raft domains, in which sphingolipids and cholesterol are highly concentrated, are believed to concentrate signaling molecules in the plasma membrane as a result of their lower fluidity than in other areas.135) Thus, lipid rafts are currently thought to be the “hot spots” for signal transduction in cells other than neurons. However, because the neuron has a much higher concentration of these lipid species than other cells,136) lipid rafts are likely to be important in neuronal signaling. Some membrane proteins are highly palmitoylated,137) which causes partitioning of membrane lipids and forms lipid rafts, incorporating such proteins.138–140) A human mutation in serine palmitoyltransferase, the enzyme that catalyzes the first step of sphingolipid synthesis, causes abnormal growth cone dynamics, implicating the importance of this enzyme in neuronal development141) (Fig. 6a).
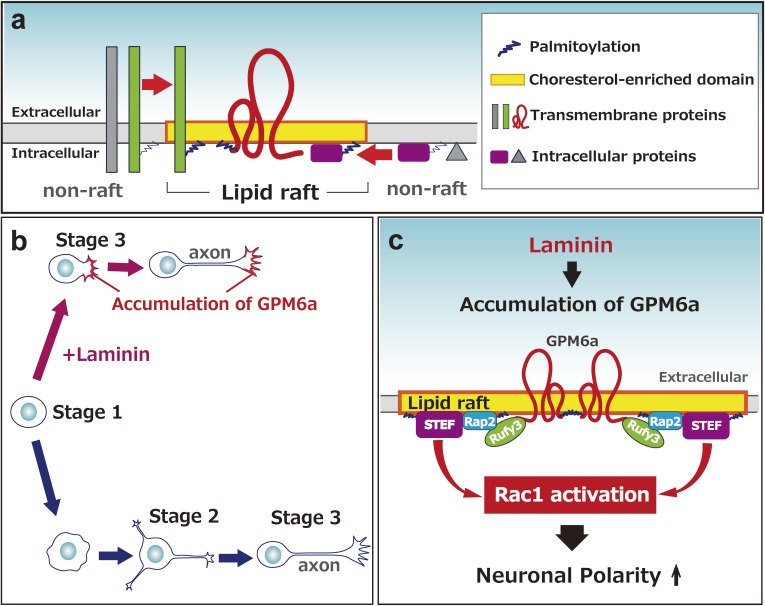
Figure 6.
Lipid rafts and palmitoylation are likely involved in neuronal development, especially in determining neuronal polarity and axon formation. (a) Lipid raft structure. Palmitoylated membrane proteins are translocated to lipid-raft domains from non-raft areas. (b) Polarity determination of neurons. Neurons are thought to have the potential for cell-autonomous polarity determination via stage 2 (multi-polar stage). However, LN- and GPM6a-dependent processes facilitate the polarity determination process by skipping stage 2 and directly moving to stage 3 (axon determination). (c) GPM6a-dependent signaling for polarity determination. Downstream proteins of GPM6a (Rufy3-Rap2-Tiam2/STEF) are gathered to lipid-raft domains in response to LN to facilitate signaling needed for neuronal polarity determination.
Our group has focused on the unique membrane protein glycoprotein, glycoprotein m6a (GPM6a), and found that lipid rafts are important for axonal growth.100,101,142) Our proteomics study indicated that GPM6a is a major membrane protein of GCM.20) GPM6a is highly expressed in differentiated neurons,143–145) although its exact function is not known. This protein has four transmembrane domains146) and is a major palmitoylated protein in the adult brain.137) We found that GPM6a is highly colocalized with cholesterol, a marker of lipid rafts.100) Additionally, lipid rafts containing GPM6a are clustered, and GPM6a itself accumulates at the origin of the neurite.
The location of growth cone formation at a specific portion of a neuron is an important unsolved problem in neuronal development. The steps in which the neuron extends processes, i.e., the axon and dendrites, during differentiation are classified into five stages in cell culture. Such an event is called neuronal polarity determination (Fig. 6b). Although the characterized cell-autonomous signaling pathways may determine neuronal polarity in individual single cells in a cell culture model system,147) as postulated by Banker and colleagues,148) according to the tug-of-war model,149) such signaling does not likely occur spontaneously and in an inconsistent way during polarization of neurons in vivo.150) In vivo, the neurons in one group should acquire their polarity simultaneously and grow their axons in the same direction. Thus, we need a new scheme that explains how extrinsic factors induce polarity determination. Two factors are known; one is the neurotrophin group,147) and the other is the extracellular matrix (ECM) component group, which includes laminin (LN).100,151) We revealed that LN facilitates neuronal polarity determination, which appears to skip stage 2.100) Such LN-induced polarity is dependent upon palmitoylated GPM6a, which is concentrated in lipid rafts. Without palmitoylation, GPM6a is localized in the plasma membrane but not in lipid rafts. As a result, neither its biased distribution at the initial growing site nor facilitation of polarity determination is induced.100)
As mentioned above, we demonstrated that palmitoylated GPM6a clusters in lipid rafts and its biased distribution is consistent with the origin site of the first neurite, in response to LN.100,101) Its downstream signaling proteins, Rufy3 (also called Singar1152)), Rap2, and Tiam2/STEF, each of which is involved in cellular polarization, are concentrated in lipid rafts in a GPM6a-dependent manner. In utero administration of RNAi against GPM6a impairs neuronal polarity in pups.100) These results suggested that GPM6a induces the clustering of downstream signaling molecules in the rafts for more rapid determination of neuronal polarity and axon formation, in response to extracellular signals100) (Fig. 6c).
Because the growth cone has lower levels of cholesterol and sphingolipids than adult synaptic membranes153) and the lipid species that compose the lipid rafts are present in much lower amounts during development than in the mature brain,154) the enrichment of signaling molecules in lipid rafts is likely to be important for the function of growth cones.
The driving of intrinsic signaling pathways such as GPM6a and its downstream signaling proteins that are concentrated in lipid rafts is likely to be important for rapid and synchronized polarity determination,147,151,155,156) which may explain why lipid rafts are thought to be important for axonal growth.157) LN probably stimulates the lipid-raft domains to cluster themselves and to increase these areas via GPM6a,100) as other guidance molecules seem to do.158,159)
We also succeeded in visualizing lipid rafts using a labeled cholesterol-binding protein and superresolution microscopy.132) The most important results, which were unexpected, were that GPM6a rescued its KO neurons in polarity determination, but the mutant-deficient palmitoylation site had no rescue activity, even though they were sufficiently expressed in the membrane.
GPM6a, Rufy3, Rap2, and Tiam1 are related to important neuropsychiatric diseases.160–163) In particular, mutations in genes for GPM6a and Rufy3 are associated with a high risk for human schizophrenia and depression, respectively. These results suggest that GPM6a and downstream molecules play physiological roles in neuronal development. GPM6a in lipid rafts is likely involved in a key step of neuronal morphogenesis.
Unsolved: GPM6a-KO and GPM-6a/6b-double KO mice have fewer abnormalities in the connections in vivo than expected from cellular studies.164) Other membrane proteins with different primary structures from the proteolipid protein (PLP) family, but which are concentrated in lipid rafts, probably compensate for GPM6a loss in the developing brain. The functions of Rufy3 seem to be various and operate via more complicated molecular interactions.165–167) However, these functions are not well understood, because the Rufy3-KO is lethal just after birth without large CNS abnormalities.101)
7. Axon regeneration via growth cone movement in vivo is regulated by chondroitin sulfate
The ECM is tightly involved in axon growth and guidance at developmental stages and successful axon regeneration. The ECM component protein, LN, is a major positive regulator of axon growth via growth cone signaling. In addition to LN, other key regulators of axon growth, including glycosaminoglycans, which are long-repeated disaccharides covalently linked to specific core proteins, are localized in the ECM. Chondroitin sulfate (CS) and heparin sulfate (HS) have related structures, but they play considerably distinct roles in growth cone behavior, i.e., CS and HS have an inhibitory and a stimulatory effect on axon growth, respectively.168) The physiological effects of CS/HS are not well characterized because good model mice for them are not available. However, some recent investigations have revealed that, for example, HS plays an important role in synaptogenesis via other membrane proteins.169,170)
We have also studied CSGalNAcT1-KO (T1KO) mice, which lack the rate-limiting enzyme for CS synthesis.121,171) In the adult brain, CS is enriched in perineuronal nets, which surround inhibitory synapses, regulate the activity of GABAergic interneurons (so-called “parvalbumin neurons”), and are involved in GABAergic synaptic plasticity.172) T1KO mice show microscopic abnormalities in the CNS including perineuronal net insufficiency and several behavioral abnormalities,173–175) but they are fertile and viable171,176) with some connective tissue abnormalities.177)
Because CS is the most abundant and potent inhibitor of axon growth and regeneration,178,179) we investigated axon regeneration after spinal cord injury (SCI) in T1KO mice. CS is synthesized by reactive astrocytes,180) which act bidirectionally on axon regeneration because they block the leakage of macrophages, which clear debris after injury, but are also toxic to the normal remaining axons. Previously, almost all studies investigating the roles of CS in SCI used bacterial chondroitinase ABC, an enzyme that degrades CS.181) However, CS is necessary in injured neural tissues and, complete loss of CS by chondroitinase ABC does not induce only “beneficial” effects. The inhibitory effects of CS on the growth cone are thought to include several signaling pathways.177,182) Axonal regeneration in SCI is inhibited at least via a CS receptor, the receptor tyrosine phosphatase sigma.183–185)
Because a reduction in CS synthesis was expected in T1KO mice, we examined recovery after SCI.186) Values of the Basso mouse score, which is used to evaluate motor function recovery after SCI in mice,187) reached as high as 7 after 8 weeks in T1KO mice.186) To the best of our knowledge, this score in T1KO mice is the highest in mice deficient in a single gene. Histologically, 40-fold more serotonin-positive motor fibers were confirmed in regions distal to the injury compared to wild-type mice.186)
We suspected that another factor plays a role in enhancing axon regeneration in addition to the reduction in CS. We focused on another glycosaminoglycan, HS, which shares a synthetic pathway with CS, just before the T1 reaction.188) The effects of HS on neuronal growth are totally opposite to those of CS.184,189–191) After SCI in T1KO mice, all HS synthesizing enzymes were rapidly upregulated, and the amounts of HS increased by more than 40-fold. Degradation of HS by bacterial heparitinase in injured areas cancelled out the positive effects in T1KO mice. Thus, we concluded that not only a decrease in CS but also upregulation of HS contributes to the marked axon regrowth in T1KO mice. These results suggested that the CS/HS ratio is essential for establishing the most suitable extracellular environment to support axon regeneration184) (Fig. 7). Including our idea, strategies for regulating CS have been developed to stimulate axonal regeneration after SCI.179)
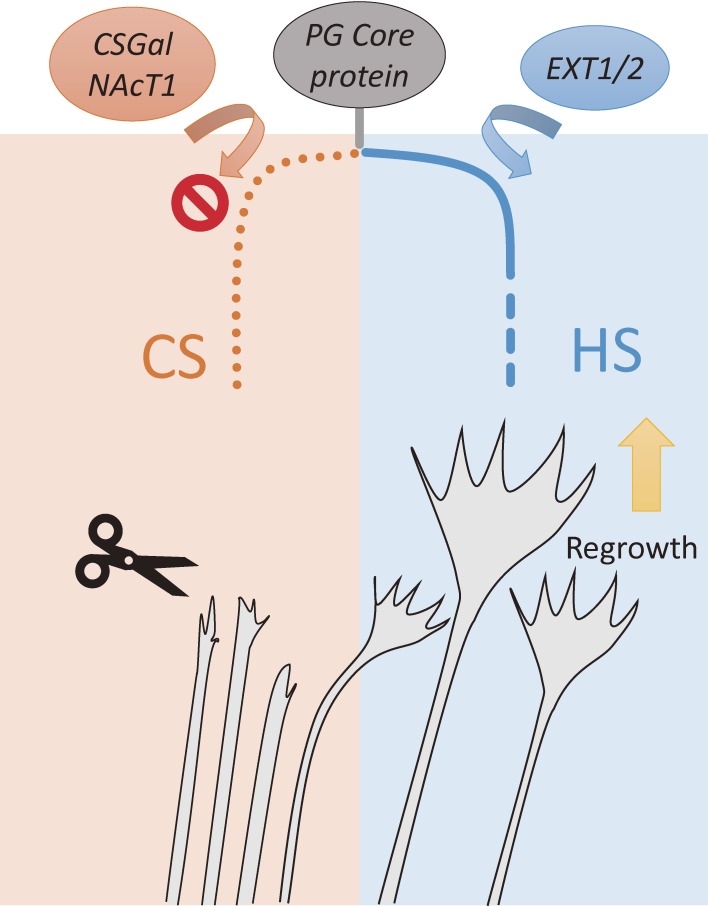
Figure 7.
Mechanisms of axon regrowth after SCI in T1KO mice. T1KO mice are deficient in T1 expression, which is normally upregulated within 24 h after SCI. CS expression is downregulated by 30%. Simultaneously, HS expression is highly upregulated by 20-fold, which is favorable for axonal growth. Inhibition of the growth cone is reduced, and stimulation of growth is elevated, respectively, and tremendous recovery from SCI occurs.
Intrinsic inhibitory mechanisms for limiting the maintenance of growth cone activity are present in the adult brain, and approaches for removing these factors have led to partially successful axon regeneration.192,193) Such mechanisms are generally energy-consuming, and maintaining them at an adult stage will be disadvantageous for energy economics in the mammalian brain, which has a large number of neurons.
Unsolved: For efficient axon regeneration after SCI, we need to elucidate the mechanisms of transcriptional regulation of glycosyltransferases, and probably sulfotransferases, which are involved in CS/HS synthesis and are thought to be the key processes. Unfortunately, neither the transcription factors nor the promoter structures for these genes are currently characterized, probably because these genes are not believed to be dynamically regulated. However, T1 is usually highly induced within 12 h of SCI,186) indicating that the mechanism of CS synthesis should be clarified. CS proteoglycans are involved in psychiatric diseases,194,195) and pathogenesis involving nerve growth and synaptogenesis should also be investigated.
8. Conclusions and future perspectives
The following points are important, unsolved problems that are essential for a complete view of the mammalian molecular mechanisms of CNS nerve growth. Most are derived from our new results using novel techniques.
8.1. 3D imaging of each component.
Our superresolution imaging revealed the importance of movements in the z-axis direction. Most membrane proteins, including guidance receptors, F-actin, and MTs, likely move three-dimensionally for axonal growth. Testing of most current molecular hypotheses of growth cone behaviors has relied on the results of 2D analysis. 3D analysis may lead to revision of the conclusions.
8.2. The sites of vesicular fusion in the growth cone.
Precisely speaking, the vesicular fusion site for axonal growth is not limited to the growth cone area; however, this is the most likely site. Why massive fusion sites have not been identified yet is one of the most important questions. Although some previous reports have described vesicular fusion sites using TIRF microscopy,196,197) only a portion of fusion sites for continuous nerve growth have likely been identified. We have partially clarified F-actin-vesicle interactions,132) but this is only one clue of the complex cytoskeletal-membrane relationship in nerve growth.198–200)
8.3. F-actin-MT interactions.
F-actin is thought to lead the growing and sliding of newly polymerizing MTs into a growth cone when the axon grows. The molecules involved in this process are not clearly understood. As described above, drebrin, MAPs, or MACF13,15) have been reported to be involved, but other linkage proteins may exist.
8.4. Cortical F-actin dynamics.
The morphology of the growth cone changes to the next, distal axon shaft when the axon grows by sliding of MTs, and this process is dependent upon cortical F-actin dynamics in the C-domain; however, this process is totally unanalyzed. Because membrane skeletal proteins, such as spectrin, ankyrin, and band 4.1 proteins, are enriched in the growth cone,15) superresolution analysis of the dynamics of these proteins will be a breakthrough in solving this problem.
8.5. Metabolism and signaling.
The growth cone should have features of metabolism due to the presence of metabolic enzymes as revealed by proteomics. However, no information has been reported concerning the relationship between metabolism and nerve growth signaling.
8.6. Retrograde signaling for nerve growth.
Growth cone signaling provides feedback to the cell body, and continuous anterograde axonal transport is needed. However, these processes are mostly unknown in the mammalian growth cone.
8.7. G-protein signaling.
Heterotrimeric G-proteins are enriched in the growth cone, as demonstrated by proteomics, but this has been known for more than a quarter of a century. Because the amounts and species of GPCRs are quite few in the growth cone,13) how G-proteins function in the growth cone is not known.
8.8. Lipid raft signaling.
Theoretically, most guidance signaling proteins should be concentrated in lipid rafts, as described above. However, this concept has not been demonstrated directly. Both more efficient systems for visualizing lipid rafts and purer fractionation of lipid raft components should be devised.
8.9. GAP-43.
This protein has been studied for more than 40 years, but its roles in the growth cone have not been clarified. Because of the considerable abundance of this protein in growing/regenerating axons and the growth cone, this protein requires additional studies.
8.10. JNK activity.
We have shown that JNK activity is indispensable for nerve growth in mammalian CNS neurons. Why and how this protein kinase is activated at developing stages are the next problems to be solved in brain development.
Acknowledgments
I thank all of the collaborators for their contributions to this work, particularly our lab members. This work was partially supported by KAKENHI from the MEXT and from JSPS.
Profile
Michihiro Igarashi was born in 1958 in Tokyo and graduated from The University of Tokyo, Faculty of Medicine (M.D.) in 1983. In 1987, he received his PhD from The University of Tokyo. He first became an assistant professor in the Department of Biochemistry at Jichi Medical School, before becoming an assistant professor and eventually an associate professor at Gunma University School of Medicine. From 1991 to 1993, he was a research fellow of medicine in the Cardiovascular Research Center at Massachusetts General Hospital-East (Charlestown, MA, U.S.A.). Since 2000, he has been a Professor and the Chair of the Department of Neurochemistry and Molecular Cell Biology, Niigata University School of Medicine and Graduate School of Medical and Dental Sciences. He was also the Vice Dean of Niigata University School of Medicine from 2010 to 2014. He was a visiting Professor at the Protein Institute of Osaka University. He was a recipient of the Niigata Nippo Newspaper Cultural Prize (2014).
References
- 1).Cajal R.S. (1890) A quelle époque apparaissent les expansions des cellules nerveuses de la moëlle épinière du poulet? Anat. Anz. 5, 609–613, 621–631. [Google Scholar]
- 2).Harrison J.G. (1910) The outgrowth of the nerve fiber as a mode of protoplasmic movement. J. Exp. Zool. 9, 787–848. [DOI] [PubMed] [Google Scholar]
- 3).Nakai J., Kawasaki Y. (1959) Studies on the mechanisms determining the course of the nerve fibers in tissue culture. I. The reaction of the growth cone to various obstructions. Z. Zellforsch. Mikrosk. Anat. 51, 108–122. [DOI] [PubMed] [Google Scholar]
- 4).Bray D. (1970) Surface movements during the growth of single explanted neurons. Proc. Natl. Acad. Sci. U.S.A. 65, 905–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5).Goodman C.S. (1996) Mechanisms and molecules that control growth cone guidance. Annu. Rev. Neurosci. 19, 341–377. [DOI] [PubMed] [Google Scholar]
- 6).Kolodkin A.L., Tessier-Lavigne M. (2011) Mechanisms and molecules of neuronal wiring: A primer. Cold Spring Harb. Perspect. Biol. 3, a001727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7).Amin L., Ercolini E., Ban J., Torre V. (2013) Comparison of the force exerted by hippocampal and DRG growth cones. PLoS One 8, e7302.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8).Dailey M.E., Bridgman P.C. (1993) Vacuole dynamics in growth cones: Correlated EM and video observations. J. Neurosci. 13, 3375–3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9).Pfenninger K.H. (2009) Plasma membrane expansion: A neuron’s Herculean task. Nat. Rev. Neurosci. 10, 251–261. [DOI] [PubMed] [Google Scholar]
- 10).Falk J., Konopacki F.A., Zivraj K.H., Holt C.E. (2014) Rab5 and rab4 regulate axon elongation in the Xenopus visual system. J. Neurosci. 34, 373–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11).Boyer N.P., Gupton S.L. (2018) Revisiting netrin-1: One who guides (axons). Front. Cell. Neurosci. 12, 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12).Sutherland D.J., Goodhill G.J. (2015) The interdependent roles of Ca2+ and cAMP in axon guidance. Dev. Neurobiol. 75, 402–410. [DOI] [PubMed] [Google Scholar]
- 13).Igarashi M. (2014) Proteomic identification of the molecular basis of mammalian CNS growth cones. Neurosci. Res. 88, 1–15. [DOI] [PubMed] [Google Scholar]
- 14).Russell S.A., Bashaw G.J. (2018) Axon guidance pathways and the control of gene expression. Dev. Dyn. 247, 571–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15).Cheng T.P., Reese T.S. (1987) Recycling of plasmalemma in chick tectal growth cones. J. Neurosci. 7, 1752–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16).Rees R.P., Bunge M.B., Bunge R.P. (1976) Morphological changes in the neuritic growth cone and target neuron during synaptic junction development in culture. J. Cell Biol. 68, 240–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17).Cheng T.P., Reese T.S. (1985) Polarized compartmentalization of organelles in growth cones from developing optic tectum. J. Cell Biol. 101, 1473–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18).Schaefer A.W., Kabir N., Forscher P. (2002) Filopodia and actin arcs guide the assembly and transport of two populations of microtubules with unique dynamic parameters in neuronal growth cones. J. Cell Biol. 158, 139–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19).Lowery L.A., Van Vactor D. (2009) The trip of the tip: Understanding the growth cone machinery. Nat. Rev. Mol. Cell Biol. 10, 332–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20).Nozumi M., Togano T., Takahashi-Niki K., Lu J., Honda A., Taoka M., et al. (2009) Identification of functional marker proteins in the mammalian growth cone. Proc. Natl. Acad. Sci. U.S.A. 106, 17211–17216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21).Jin L.Q., Pennise C.R., Rodemer W., Jahn K.S., Selzer M.E. (2016) Protein synthetic machinery and mRNA in regenerating tips of spinal cord axons in lamprey. J. Comp. Neurol. 524, 3614–3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22).Fukura H., Komiya Y., Igarashi M. (1996) Signaling pathway downstream of GABAA receptor in the growth cone. J. Neurochem. 67, 1426–1434. [DOI] [PubMed] [Google Scholar]
- 23).Zheng J.Q., Poo M.M. (2007) Calcium signaling in neuronal motility. Annu. Rev. Cell Dev. Biol. 23, 375–404. [DOI] [PubMed] [Google Scholar]
- 24).Mortimer D., Fothergill T., Pujic Z., Richards L.J., Goodhill G.J. (2008) Growth cone chemotaxis. Trends Neurosci. 31, 90–98. [DOI] [PubMed] [Google Scholar]
- 25).Tojima T., Hines J.H., Henley J.R., Kamiguchi H. (2011) Second messengers and membrane trafficking direct and organize growth cone steering. Nat. Rev. Neurosci. 12, 191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26).Pignata A., Ducuing H., Castellani V. (2016) Commissural axon navigation: Control of midline crossing in the vertebrate spinal cord by the semaphorin 3B signaling. Cell Adhes. Migr. 10, 604–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27).Stoeckli E.T. (2018) Understanding axon guidance: Are we nearly there yet? Development 145, dev151415. [DOI] [PubMed] [Google Scholar]
- 28).Craft G.E., Chen A., Nairn A.C. (2013) Recent advances in quantitative neuroproteomics. Methods 61, 186–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29).Pfenninger K.H., Ellis L., Johnson M.P., Friedman L.B., Somlo S. (1983) Nerve growth cones isolated from fetal rat brain: Subcellular fractionation and characterization. Cell 35, 573–584. [DOI] [PubMed] [Google Scholar]
- 30).Gordon-Weeks P.R., Lockerbie R.O. (1984) Isolation and partial characterisation of neuronal growth cones from neonatal rat forebrain. Neuroscience 13, 119–136. [DOI] [PubMed] [Google Scholar]
- 31).Ellis L., Wallis I., Abreu E., Pfenninger K.H. (1985) Nerve growth cones isolated from fetal rat brain. IV. Preparation of a membrane subfraction and identification of a membrane glycoprotein expressed on sprouting neurons. J. Cell Biol. 101, 1977–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32).Gordon-Weeks P.R. (1988) The ultrastructure of the neuronal growth cone: New insights from subcellular fractionation and rapid freezing studies. Electron Microsc. Rev. 1, 201–219. [DOI] [PubMed] [Google Scholar]
- 33).Estrada-Bernal A., Sanford S.D., Sosa L.S., Simon G.C., Hansen K.C., Pfenninger K.H. (2012) Functional complexity of the axonal growth cone: A proteomic analysis. PLoS One 7, e31858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34).Bai F., Witzmann F.A. (2007) Synaptosome proteomics. Subcell. Biochem. 43, 77–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35).Igarashi, M. and Ohko, K. (2009) Proteins involved in the presynaptic functions. In Handbook of Neurochemistry and Molecular Neurobiology: Neural Signaling Mecahmism, 3rd ed. (eds. Lajtha, A. and Mikoshiba, K.). Springer, New York, pp. 47–62. [Google Scholar]
- 36).Südhof T.C. (2013) Neurotransmitter release: The last millisecond in the life of a synaptic vesicle. Neuron 80, 675–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37).Denny J.B. (2006) Molecular mechanisms, biological actions, and neuropharmacology of the growth-associated protein GAP-43. Curr. Neuropharmacol. 4, 293–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38).Lu J., Nozumi M., Fuji H., Igarashi M. (2008) A novel method for RNA interference in neurons using enhanced green fluorescent protein (EGFP)-transgenic rats. Neurosci. Res. 61, 219–224. [DOI] [PubMed] [Google Scholar]
- 39).Frese C.K., Mikhaylova M., Stucchi R., Gautier V., Liu Q., Mohammed S., et al. (2017) Quantitative map of proteome dynamics during neuronal differentiation. Cell Rep. 18, 1527–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40).Cagnetta R., Frese C.K., Shigeoka T., Krijgsveld J., Holt C.E. (2018) Rapid cue-specific remodeling of the nascent axonal proteome. Neuron 99, 29–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41).Toriyama M., Shimada T., Kim K.B., Mitsuba M., Nomura E., Katsuta K., et al. (2006) Shootin1: A protein involved in the organization of an asymmetric signal for neuronal polarization. J. Cell Biol. 175, 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42).Tortosa E., Adolfs Y., Fukata M., Pasterkamp R.J., Kapitein L.C., Hoogenraad C.C. (2017) Dynamic palmitoylation targets map6 to the axon to promote microtubule stabilization during neuronal polarization. Neuron 94, 809–826. [DOI] [PubMed] [Google Scholar]
- 43).Deloulme J.C., Gory-Fauré S., Mauconduit F., Chauvet S., Jonckheere J., Boulan B., et al. (2015) Microtubule-associated protein 6 mediates neuronal connectivity through Semaphorin 3E-dependent signalling for axonal growth. Nat. Commun. 6, 7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44).Akhmanova A., Steinmetz M.O. (2010) Microtubule +TIPs at a glance. J. Cell Sci. 123, 3415–3419. [DOI] [PubMed] [Google Scholar]
- 45).Cammarata G.M., Bearce E.A., Lowery L.A. (2016) Cytoskeletal social networking in the growth cone: How +TIPs mediate microtubule-actin cross-linking to drive axon outgrowth and guidance. Cytoskeleton (Hoboken) 73, 461–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46).Nwagbara B.U., Faris A.E., Bearce E.A., Erdogan B., Ebbert P.T., Evans M.F., et al. (2014) TACC3 is a microtubule plus end-tracking protein that promotes axon elongation and also regulates microtubule plus end dynamics in multiple embryonic cell types. Mol. Biol. Cell 25, 3350–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47).Dent E.W., Gupton S.L., Gertler F.B. (2011) The growth cone cytoskeleton in axon outgrowth and guidance. Cold Spring Harb. Perspect. Biol. 3, a001800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48).Omotade O.F., Pollitt S.L., Zheng J.Q. (2017) Actin-based growth cone motility and guidance. Mol. Cell. Neurosci. 84, 4–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49).Moradi M., Sivadasan R., Saal L., Lüningschrör P., Dombert B., Rathod R.J., et al. (2017) Differential roles of α-, β-, and γ-actin in axon growth and collateral branch formation in motoneurons. J. Cell Biol. 216, 793–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50).Tsygankov D., Bilancia C.G., Vitriol E.A., Hahn K.M., Peifer M., Elston T.C. (2014) CellGeo: A computational platform for the analysis of shape changes in cells with complex geometries. J. Cell Biol. 204, 443–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51).Urbančič V., Butler R., Richier B., Peter M., Mason J., Livesey F.J., et al. (2017) Filopodyan: An open-source pipeline for the analysis of filopodia. J. Cell Biol. 216, 3405–3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52).Tamada A., Igarashi M. (2017) Revealing chiral cell motility by 3D Riesz transform-differential interference contrast microscopy and computational kinematic analysis. Nat. Commun. 8, 2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53).Bagonis M.M., Fusco L., Pertz O., Danuser G. (2019) Automated profiling of growth cone heterogeneity defines relations between morphology and motility. J. Cell Biol. 218, 350–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54).Iuliano O., Yoshimura A., Prospéri M.T., Martin R., Knölker H.J., Coudrier E. (2018) Myosin 1b promotes axon formation by regulating actin wave propagation and growth cone dynamics. J. Cell Biol. 217, 2033–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55).Katsuno H., Toriyama M., Hosokawa Y., Mizuno K., Ikeda K., Sakumura Y., et al. (2015) Actin migration driven by directional assembly and disassembly of membrane-anchored actin filaments. Cell Rep. 12, 648–660. [DOI] [PubMed] [Google Scholar]
- 56).Mortal S., Iseppon F., Perissinotto A., D’Este E., Cojoc D., Napolitano L.M.R., et al. (2017) Actin waves do not boost neurite outgrowth in the early stages of neuron maturation. Front. Cell. Neurosci. 11, 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57).Kamijo S., Ishii Y., Horigane S.I., Suzuki K., Ohkura M., Nakai J., et al. (2018) A critical neurodevelopmental role for L-type voltage-gated calcium channels in neurite extension and radial migration. J. Neurosci. 38, 5551–5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58).Meiri K.F., Gordon-Weeks P.R. (1990) GAP-43 in growth cones is associated with areas of membrane that are tightly bound to substrate and is a component of a membrane skeleton subcellular fraction. J. Neurosci. 10, 256–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59).Strittmatter S.M., Valenzuela D., Kennedy T.E., Neer E.J., Fishman M.C. (1990) GO is a major growth cone protein subject to regulation by GAP-43. Nature 344, 836–841. [DOI] [PubMed] [Google Scholar]
- 60).Igarashi M., Strittmatter S.M., Vartanian T., Fishman M.C. (1993) Mediation by G proteins of signals that cause collapse of growth cones. Science 259, 77–79. [DOI] [PubMed] [Google Scholar]
- 61).Saito S., Komiya Y., Igarashi M. (1991) Muscarinic acetylcholine receptors are expressed and enriched in growth cone membranes isolated from fetal and neonatal rat forebrain: Pharmacological demonstration and characterization. Neuroscience 45, 735–745. [DOI] [PubMed] [Google Scholar]
- 62).Mauriac S.A., Hien Y.E., Bird J.E., Carvalho S.D., Peyroutou R., Lee S.C., et al. (2017) Defective Gpsm2/Gαi3 signalling disrupts stereocilia development and growth cone actin dynamics in Chudley-McCullough syndrome. Nat. Commun. 8, 14907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63).Vysokov N.V., Silva J.P., Lelianova V.G., Suckling J., Cassidy J., Blackburn J.K., et al. (2018) Proteolytically released Lasso/teneurin-2 induces axonal attraction by interacting with latrophilin-1 on axonal growth cones. eLife 7, e37935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64).Woelfle R., D’Aquila A.L., Lovejoy D.A. (2016) Teneurins, TCAP, and latrophilins: Roles in the etiology of mood disorders. Transl. Neurosci. 7, 17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65).Wang T., Liu Y., Xu X.H., Deng C.Y., Wu K.Y., Zhu J., et al. (2011) Lgl1 activation of rab10 promotes axonal membrane trafficking underlying neuronal polarization. Dev. Cell 21, 431–444. [DOI] [PubMed] [Google Scholar]
- 66).Nakazawa H., Sada T., Toriyama M., Tago K., Sugiura T., Fukuda M., et al. (2012) Rab33a mediates anterograde vesicular transport for membrane exocytosis and axon outgrowth. J. Neurosci. 32, 12712–12725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67).Villarroel-Campos D., Bronfman F.C., Gonzalez-Billault C. (2016) Rab GTPase signaling in neurite outgrowth and axon specification. Cytoskeleton (Hoboken) 73, 498–507. [DOI] [PubMed] [Google Scholar]
- 68).Koseki H., Donegá M., Lam B.Y., Petrova V., van Erp S., Yeo G.S., et al. (2017) Selective rab11 transport and the intrinsic regenerative ability of CNS axons. eLife 6, e26956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69).Inagaki N., Chihara K., Arimura N., Ménager C., Kawano Y., Matsuo N., et al. (2001) CRMP-2 induces axons in cultured hippocampal neurons. Nat. Neurosci. 4, 781–782. [DOI] [PubMed] [Google Scholar]
- 70).Khazaei M.R., Girouard M.P., Alchini R., Ong Tone S., Shimada T., Bechstedt S., et al. (2014) Collapsin response mediator protein 4 regulates growth cone dynamics through the actin and microtubule cytoskeleton. J. Biol. Chem. 289, 30133–30143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71).Tan M., Cha C., Ye Y., Zhang J., Li S., Wu F., et al. (2015) CRMP4 and CRMP2 interact to coordinate cytoskeleton dynamics, regulating growth cone development and axon elongation. Neural Plast. 2015, 947423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72).Kawano Y., Yoshimura T., Tsuboi D., Kawabata S., Kaneko-Kawano T., Shirataki H., et al. (2005) CRMP-2 is involved in kinesin-1-dependent transport of the Sra-1/WAVE1 complex and axon formation. Mol. Cell. Biol. 25, 9920–9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73).Bustos D.M. (2012) The role of protein disorder in the 14-3-3 interaction network. Mol. Biosyst. 8, 178–184. [DOI] [PubMed] [Google Scholar]
- 74).Cornell B., Toyo-Oka K. (2017) 14-3-3 Proteins in brain development: Neurogenesis, neuronal migration and neuromorphogenesis. Front. Mol. Neurosci. 10, 318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75).Kaplan A., Morquette B., Kroner A., Leong S., Madwar C., Sanz R., et al. (2017) Small-molecule stabilization of 14-3-3 protein-protein interactions stimulates axon regeneration. Neuron 93, 1082–1094. [DOI] [PubMed] [Google Scholar]
- 76).Campbell D.S., Holt C.E. (2001) Chemotropic responses of retinal growth cones mediated by rapid local protein synthesis and degradation. Neuron 32, 1013–1026. [DOI] [PubMed] [Google Scholar]
- 77).Menon S., Boyer N.P., Winkle C.C., McClain L.M., Hanlin C.C., Pandey D., et al. (2015) The E3 ubiquitin ligase TRIM9 is a filopodia off switch required for netrin-dependent axon guidance. Dev. Cell 35, 698–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78).van Beuningen S.F.B., Will L., Harterink M., Chazeau A., van Battum E.Y., Frias C.P., et al. (2015) TRIM46 controls neuronal polarity and axon specification by driving the formation of parallel microtubule arrays. Neuron 88, 1208–1226. [DOI] [PubMed] [Google Scholar]
- 79).Osen-Sand A., Catsicas M., Staple J.K., Jones K.A., Ayala G., Knowles J., et al. (1993) Inhibition of axonal growth by SNAP-25 antisense oligonucleotides in vitro and in vivo. Nature 364, 445–448. [DOI] [PubMed] [Google Scholar]
- 80).Igarashi M., Kozaki S., Terakawa S., Kawano S., Ide C., Komiya Y. (1996) Growth cone collapse and inhibition of neurite growth by Botulinum neurotoxin C1: A t-SNARE is involved in axonal growth. J. Cell Biol. 134, 205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81).Igarashi M., Tagaya M., Komiya Y. (1997) The soluble N-ethylmaleimide-sensitive factor attached (SNARE) protein receptor complex in growth cones: Molecular aspects of the axon terminal development. J. Neurosci. 17, 1460–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82).Kabayama H., Takeuchi M., Taniguchi M., Tokushige N., Kozaki S., Mizutani A., et al. (2011) Syntaxin 1B suppresses macropinocytosis and semaphorin 3A-induced growth cone collapse. J. Neurosci. 31, 7357–7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83).Akiyama H., Fukuda T., Tojima T., Nikolaev V.O., Kamiguchi H. (2016) Cyclic nucleotide control of microtubule dynamics for axon guidance. J. Neurosci. 36, 5636–5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84).Barrecheguren P.J., Ros O., Cotrufo T., Kunz B., Soriano E., Ulloa F., et al. (2017) SNARE proteins play a role in motor axon guidance in vertebrates and invertebrates. Dev. Neurobiol. 77, 963–974. [DOI] [PubMed] [Google Scholar]
- 85).Ros O., Barrecheguren P.J., Cotrufo T., Schaettin M., Roselló-Busquets C., Vílchez-Acosta A., et al. (2018) A conserved role for Syntaxin-1 in pre- and post-commissural midline axonal guidance in fly, chick, and mouse. PLoS Genet. 14, e1007432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86).Fuschini G., Cotrufo T., Ros O., Muhaisen A., Andrés R., Comella J.X., et al. (2018) Syntaxin-1/TI-VAMP SNAREs interact with Trk receptors and are required for neurotrophin-dependent outgrowth. Oncotarget 9, 35922–35940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87).Chen L., Liu Z., Zhou B., Wei C., Zhou Y., Rosenfeld M.G., et al. (2016) CELF RNA binding proteins promote axon regeneration in C. elegans and mammals through alternative splicing of Syntaxins. eLife 5, e16072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88).Humphrey S.J., James D.E., Mann M. (2015) Protein phosphorylation: A major switch mechanism for metabolic regulation. Trends Endocrinol. Metab. 26, 676–687. [DOI] [PubMed] [Google Scholar]
- 89).Lisman J., Yasuda R., Raghavachari S. (2012) Mechanisms of CaMKII action in long-term potentiation. Nat. Rev. Neurosci. 13, 169–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90).Mosevitsky M.I. (2005) Nerve ending “signal” proteins GAP-43, MARCKS, and BASP1. Int. Rev. Cytol. 245, 245–325. [DOI] [PubMed] [Google Scholar]
- 91).Radu M., Chernoff J. (2017) Recent advances in methods to assess the activity of the kinome. F1000Res. 6, 1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92).Liao L., McClatchy D.B., Yates J.R. (2009) Shotgun proteomics in neuroscience. Neuron 63, 12–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93).von Stechow L., Francavilla C., Olsen J.V. (2015) Recent findings and technological advances in phosphoproteomics for cells and tissues. Expert Rev. Proteomics 12, 469–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94).Invergo B.M., Beltrao P. (2018) Reconstructing phosphorylation signalling networks from quantitative phosphoproteomic data. Essays Biochem. 62, 525–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95).Kawasaki A., Okada M., Tamada A., Okuda S., Nozumi M., Ito Y., et al. (2018) Growth cone phosphoproteomics reveals that GAP-43 phosphorylated by JNK is a marker of axon growth and regeneration. Science 4, 190–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96).Villén J., Beausoleil S.A., Gerber S.A., Gygi S.P. (2007) Large-scale phosphorylation analysis of mouse liver. Proc. Natl. Acad. Sci. U.S.A. 104, 1488–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97).Bogoyevitch M.A., Ngoei K.R., Zhao T.T., Yeap Y.Y., Ng D.C. (2010) c-Jun N-terminal kinase (JNK) signaling: Recent advances and challenges. Biochim. Biophys. Acta 1804, 463–475. [DOI] [PubMed] [Google Scholar]
- 98).Shin J.E., Miller B.R., Babetto E., Cho Y., Sasaki Y., Qayum S., et al. (2012) SCG10 is a JNK target in the axonal degeneration pathway. Proc. Natl. Acad. Sci. U.S.A. 109, E3696–E3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99).Tararuk T., Ostman N., Li W., Björkblom B., Padzik A., Zdrojewska J., et al. (2006) JNK1 phosphorylation of SCG10 determines microtubule dynamics and axodendritic length. J. Cell Biol. 173, 265–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100).Honda A., Ito Y., Takahashi-Niki K., Matsushita N., Nozumi M., Tabata H., et al. (2017) Extracellular signals induce glycoprotein M6a clustering of lipid rafts and associated signaling molecules. J. Neurosci. 37, 4046–4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101).Honda A., Usui H., Sakimura K., Igarashi M. (2017) Rufy3 is an adapter protein for small GTPases that activates a Rac guanine nucleotide exchange factor to control neuronal polarity. J. Biol. Chem. 292, 20936–20946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102).Hirai S., Cui D.F., Miyata T., Ogawa M., Kiyonari H., Suda Y., et al. (2006) The c-Jun N-terminal kinase activator dual leucine zipper kinase regulates axon growth and neuronal migration in the developing cerebral cortex. J. Neurosci. 26, 11992–12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103).Hirai S., Banba Y., Satake T., Ohno S. (2011) Axon formation in neocortical neurons depends on stage-specific regulation of microtubule stability by the dual leucine zipper kinase-c-Jun N-terminal kinase pathway. J. Neurosci. 31, 6468–6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104).Yamasaki T., Kawasaki H., Nishina H. (2012) Diverse roles of JNK and MKK pathways in the brain. J. Signal Transduct. 2012, 459265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105).Yamasaki T., Deki-Arima N., Kaneko A., Miyamura N., Iwatsuki M., Matsuoka M., et al. (2017) Age-dependent motor dysfunction due to neuron-specific disruption of stress-activated protein kinase MKK7. Sci. Rep. 7, 7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106).Coffey E.T. (2014) Nuclear and cytosolic JNK signalling in neurons. Nat. Rev. Neurosci. 15, 285–299. [DOI] [PubMed] [Google Scholar]
- 107).Zhang L., Zhang P., Wang G., Zhang H., Zhang Y., Yu Y., et al. (2018) Ras and rap signal bidirectional synaptic plasticity via distinct subcellular microdomains. Neuron 98, 783–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108).Hollos P., Marchisella F., Coffey E.T. (2018) JNK regulation of depression and anxiety. Br. Plast. 3, 145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109).Mohamed A., Robinson H., Erramouspe P.J., Hill M.M. (2018) Advances and challenges in understanding the role of the lipid raft proteome in human health. Expert Rev. Proteomics 15, 1053–1063. [DOI] [PubMed] [Google Scholar]
- 110).Hanz S., Fainzilber M. (2006) Retrograde signaling in injured nerve—The axon reaction revisited. J. Neurochem. 99, 13–19. [DOI] [PubMed] [Google Scholar]
- 111).Myers A.K., Meechan D.W., Adney D.R., Tucker E.S. (2014) Cortical interneurons require Jnk1 to enter and navigate the developing cerebral cortex. J. Neurosci. 34, 7787–7801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112).Qu C., Li W., Shao Q., Dwyer T., Huang H., Yang T., et al. (2013) c-Jun N-terminal kinase 1 (JNK1) is required for coordination of netrin signaling in axon guidance. J. Biol. Chem. 288, 1883–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113).Zheng M., Chen R., Chen H., Zhang Y., Chen J., Lin P., et al. (2018) Netrin-1 promotes synaptic formation and axonal regeneration via jnk1/c-jun pathway after the middle cerebral artery occlusion. Front. Cell. Neurosci. 12, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114).Skene J.H. (1989) Axonal growth-associated proteins. Annu. Rev. Neurosci. 12, 127–156. [DOI] [PubMed] [Google Scholar]
- 115).Holahan M.R. (2017) A shift from a pivotal to supporting role for the growth-associated protein (GAP-43) in the coordination of axonal structural and functional plasticity. Front. Cell. Neurosci. 11, 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116).Spencer S.A., Schuh S.M., Liu W.S., Willard M.B. (1992) GAP-43, a protein associated with axon growth, is phosphorylated at three sites in cultured neurons and rat brain. J. Biol. Chem. 267, 9059–9064. [PubMed] [Google Scholar]
- 117).Hibi M., Lin A., Smeal T., Minden A., Karin M. (1993) Identification of an oncoprotein- and UV-responsive protein kinase that binds and potentiates the c-Jun activation domain. Genes Dev. 7, 2135–2148. [DOI] [PubMed] [Google Scholar]
- 118).Oyamatsu H., Koga D., Igarashi M., Shibata M., Ushiki T. (2012) Morphological assessment of early axonal regeneration in end-to-side nerve coaptation models. J. Plast. Surg. Hand Surg. 46, 299–307. [DOI] [PubMed] [Google Scholar]
- 119).Strittmatter S.M., Fankhauser C., Huang P.L., Mashimo H., Fishman M.C. (1995) Neuronal pathfinding is abnormal in mice lacking the neuronal growth cone protein GAP-43. Cell 80, 445–452. [DOI] [PubMed] [Google Scholar]
- 120).Flamm A.G., Żerko S., Zawadzka-Kazimierczuk A., Koźmiński W., Konrat R., Coudevylle N. (2016) 1H, 15N, 13C resonance assignment of human GAP-43. Biomol. NMR Assign. 10, 171–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121).Sudo Y., Valenzuela D., Beck-Sickinger A.G., Fishman M.C., Strittmatter S.M. (1992) Palmitoylation alters protein activity: Blockade of Go stimulation by GAP-43. EMBO J. 11, 2095–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122).Igarashi M., Nozumi M., Wu L.G., Zanacchi F.C., Katona I., Barna L., et al. (2018) New observations in neuroscience using superresolution microscopy. J. Neurosci. 38, 9459–9467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123).Schermelleh L., Ferrand A., Huser T., Eggeling C., Sauer M., Biehlmaier O., et al. (2019) Super-resolution microscopy demystified. Nat. Cell Biol. 21, 72–84. [DOI] [PubMed] [Google Scholar]
- 124).Ganguly A., Tang Y., Wang L., Ladt K., Loi J., Dargent B., et al. (2015) A dynamic formin-dependent deep F-actin network in axons. J. Cell Biol. 210, 401–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125).Unsain N., Bordenave M.D., Martinez G.F., Jalil S., von Bilderling C., Barabas F.M., et al. (2018) Remodeling of the actin/spectrin membrane-associated periodic skeleton, growth cone collapse and f-actin decrease during axonal degeneration. Sci. Rep. 8, 3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126).Dubey P., Jorgenson K., Roy S. (2018) Actin assemblies in the axon shaft—Some open questions. Curr. Opin. Neurobiol. 51, 163–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127).Papandréou M.J., Leterrier C. (2018) The functional architecture of axonal actin. Mol. Cell. Neurosci. 91, 151–159. [DOI] [PubMed] [Google Scholar]
- 128).Biswas S., Kalil K. (2018) The microtubule-associated protein tau mediates the organization of microtubules and their dynamic exploration of actin-rich lamellipodia and filopodia of cortical growth cones. J. Neurosci. 38, 291–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129).Gustafsson M.G. (2000) Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J. Microsc. 198, 82–87. [DOI] [PubMed] [Google Scholar]
- 130).Demmerle J., Innocent C., North A.J., Ball G., Müller M., Miron E., et al. (2017) Strategic and practical guidelines for successful structured illumination microscopy. Nat. Protoc. 12, 988–1010. [DOI] [PubMed] [Google Scholar]
- 131).Richter K.N., Rizzoli S.O., Jähne S., Vogts A., Lovric J. (2017) Review of combined isotopic and optical nanoscopy. Neurophotonics 4, 020901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132).Nozumi M., Nakatsu F., Katoh K., Igarashi M. (2017) Coordinated movement of vesicles and actin bundles during nerve growth revealed by superresolution microscopy. Cell Rep. 18, 2203–2218. [DOI] [PubMed] [Google Scholar]
- 133).Nozumi M., Igarashi M. (2018) Vesicular movements of the growth cone. Neurochem. Int. 119, 71–76. [DOI] [PubMed] [Google Scholar]
- 134).Boucrot E., Ferreira A.P., Almeida-Souza L., Debard S., Vallis Y., Howard G., et al. (2015) Endophilin marks and controls a clathrin-independent endocytic pathway. Nature 517, 460–465. [DOI] [PubMed] [Google Scholar]
- 135).Lingwood D., Simons K. (2010) Lipid rafts as a membrane-organizing principle. Science 327, 46–50. [DOI] [PubMed] [Google Scholar]
- 136).Aureli M., Grassi S., Prioni S., Sonnino S., Prinetti A. (2015) Lipid membrane domains in the brain. Biochim. Biophys. Acta 1851, 1006–1016. [DOI] [PubMed] [Google Scholar]
- 137).Kang R., Wan J., Arstikaitis P., Takahashi H., Huang K., Bailey A.O., et al. (2008) Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature 456, 904–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138).Lorent J.H., Levental I. (2015) Structural determinants of protein partitioning into ordered membrane domains and lipid rafts. Chem. Phys. Lipids 192, 23–32. [DOI] [PubMed] [Google Scholar]
- 139).Globa A.K., Bamji S.X. (2017) Protein palmitoylation in the development and plasticity of neuronal connections. Curr. Opin. Neurobiol. 45, 210–220. [DOI] [PubMed] [Google Scholar]
- 140).Lin X., Gorfe A.A., Levental I. (2018) Protein partitioning into ordered membrane domains: Insights from simulations. Biophys. J. 114, 1936–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141).Jun B.K., Chandra A., Kuljis D., Schmidt B.P., Eichler F.S. (2015) Substrate availability of mutant SPT alters neuronal branching and growth cone dynamics in dorsal root ganglia. J. Neurosci. 35, 13713–13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142).Ito Y., Honda A., Igarashi M. (2018) Glycoprotein M6a as a signaling transducer in neuronal lipid rafts. Neurosci. Res. 128, 19–24. [DOI] [PubMed] [Google Scholar]
- 143).Yan Y., Lagenaur C., Narayanan V. (1993) Molecular cloning of M6: Identification of a PLP/DM20 gene family. Neuron 11, 423–431. [DOI] [PubMed] [Google Scholar]
- 144).Alfonso J., Fernández M.E., Cooper B., Flugge G., Frasch A.C. (2005) The stress-regulated protein M6a is a key modulator for neurite outgrowth and filopodium/spine formation. Proc. Natl. Acad. Sci. U.S.A. 102, 17196–17201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145).Michibata H., Okuno T., Konishi N., Wakimoto K., Kyono K., Aoki K., et al. (2008) Inhibition of mouse GPM6A expression leads to decreased differentiation of neurons derived from mouse embryonic stem cells. Stem Cells Dev. 17, 641–651. [DOI] [PubMed] [Google Scholar]
- 146).Möbius W., Patzig J., Nave K.A., Werner H.B. (2008) Phylogeny of proteolipid proteins: Divergence, constraints, and the evolution of novel functions in myelination and neuroprotection. Neuron Glia Biol. 4, 111–127. [DOI] [PubMed] [Google Scholar]
- 147).Takano T., Xu C., Funahashi Y., Namba T., Kaibuchi K. (2015) Neuronal polarization. Development 142, 2088–2093. [DOI] [PubMed] [Google Scholar]
- 148).Dotti C.G., Sullivan C.A., Banker G.A. (1988) The establishment of polarity by hippocampal neurons in culture. J. Neurosci. 8, 1454–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149).Lalli G. (2014) Regulation of neuronal polarity. Exp. Cell Res. 328, 267–275. [DOI] [PubMed] [Google Scholar]
- 150).Namba T., Kibe Y., Funahashi Y., Nakamuta S., Takano T., Ueno T., et al. (2014) Pioneering axons regulate neuronal polarization in the developing cerebral cortex. Neuron 81, 814–829. [DOI] [PubMed] [Google Scholar]
- 151).Esch T., Lemmon V., Banker G. (1999) Local presentation of substrate molecules directs axon specification by cultured hippocampal neurons. J. Neurosci. 19, 6417–6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152).Mori T., Wada T., Suzuki T., Kubota Y., Inagaki N. (2007) Singar1, a novel RUN domain-containing protein, suppresses formation of surplus axons for neuronal polarity. J. Biol. Chem. 282, 19884–19893. [DOI] [PubMed] [Google Scholar]
- 153).Igarashi M., Waki H., Hirota M., Hirabayashi Y., Obata K., Ando S. (1990) Differences in lipid composition between isolated growth cones from the forebrain and those from the brainstem in the fetal rat. Brain Res. Dev. Brain Res. 51, 1–9. [DOI] [PubMed] [Google Scholar]
- 154).Tulodziecka K., Diaz-Rohrer B.B., Farley M.M., Chan R.B., Di Paolo G., Levental K.R., et al. (2016) Remodeling of the postsynaptic plasma membrane during neural development. Mol. Biol. Cell 27, 3480–3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155).Ménager C., Arimura N., Fukata Y., Kaibuchi K. (2004) PIP3 is involved in neuronal polarization and axon formation. J. Neurochem. 89, 109–118. [DOI] [PubMed] [Google Scholar]
- 156).Randlett O., Poggi L., Zolessi F.R., Harris W.A. (2011) The oriented emergence of axons from retinal ganglion cells is directed by laminin contact in vivo. Neuron 70, 266–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157).Kamiguchi H. (2006) The region-specific activities of lipid rafts during axon growth and guidance. J. Neurochem. 98, 330–335. [DOI] [PubMed] [Google Scholar]
- 158).Tassew N.G., Mothe A.J., Shabanzadeh A.P., Banerjee P., Koeberle P.D., Bremner R., et al. (2014) Modifying lipid rafts promotes regeneration and functional recovery. Cell Rep. 8, 1146–1159. [DOI] [PubMed] [Google Scholar]
- 159).Averaimo S., Assali A., Ros O., Couvet S., Zagar Y., Genescu I. (2016) A plasma membrane microdomain compartmentalizes ephrin-generated cAMP signals to prune developing retinal axon arbors. Nat. Commun. 7, 12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160).Ma C., Gu C., Huo Y., Li X., Luo X.J. (2018) The integrated landscape of causal genes and pathways in schizophrenia. Transl. Psychiatry 8, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161).Aberg K.A., Dean B., Shabalin A.A., Chan R.F., Han L.K.M., Zhao M., et al. (2018) Methylome-wide association findings for major depressive disorder overlap in blood and brain and replicate in independent brain samples. Mol. Psychiatry, doi: 10.1038/s41380-018-0247-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162).Funk A.J., McCullumsmith R.E., Haroutunian V., Meador-Woodruff J.H. (2012) Abnormal activity of the MAPK- and cAMP-associated signaling pathways in frontal cortical areas in postmortem brain in schizophrenia. Neuropsychopharmacology 37, 896–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163).Bhattacherjee A., Mu Y., Winter M.K., Knapp J.R., Eggimann L.S., Gunewardena S.S., et al. (2017) Neuronal cytoskeletal gene dysregulation and mechanical hypersensitivity in a rat model of Rett syndrome. Proc. Natl. Acad. Sci. U.S.A. 114, E6952–E6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164).Mita S., de Monasterio-Schrader P., Fünfschilling U., Kawasaki T., Mizuno H., Iwasato T., et al. (2015) Transcallosal projections require glycoprotein m6-dependent neurite growth and guidance. Cereb. Cortex 25, 4111–4125. [DOI] [PubMed] [Google Scholar]
- 165).Wang G., Zhang Q., Song Y., Wang X., Guo Q., Zhang J., et al. (2015) PAK1 regulates RUFY3-mediated gastric cancer cell migration and invasion. Cell Death Dis. 6, e1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166).Zelaya M.V., Pérez-Valderrama E., de Morentin X.M., Tuñon T., Ferrer I., Luquin M.R., et al. (2015) Olfactory bulb proteome dynamics during the progression of sporadic Alzheimer’s disease: Identification of common and distinct olfactory targets across Alzheimer-related co-pathologies. Oncotarget 6, 39437–39456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167).Xie R., Wang J., Liu X., Wu L., Zhang H., Tang W., et al. (2017) RUFY3 interaction with FOXK1 promotes invasion and metastasis in colorectal cancer. Sci. Rep. 7, 3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168).Dyck S.M., Karimi-Abdolrezaee S. (2015) Chondroitin sulfate proteoglycans: Key modulators in the developing and pathologic central nervous system. Exp. Neurol. 269, 169–187. [DOI] [PubMed] [Google Scholar]
- 169).Zhang P., Lu H., Peixoto R.T., Pines M.K., Ge Y., Oku S., et al. (2018) Heparan sulfate organizes neuronal synapses through neurexin partnerships. Cell 174, 1450–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170).Condomitti G., Wierda K.D., Schroeder A., Rubio S.E., Vennekens K.M., Orlandi C., et al. (2018) An input-specific orphan receptor GPR158-HSPG interaction organizes hippocampal mossy fiber-CA3 synapses. Neuron 100, 201–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171).Watanabe Y., Takeuchi K., Higa Onaga S., Sato M., Tsujita M., Abe M., et al. (2010) Chondroitin sulfate N-acetylgalactosaminyltransferase-1 is required for normal cartilage development. Biochem. J. 432, 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172).Tsien R.Y. (2013) Very long-term memories may be stored in the pattern of holes in the perineuronal net. Proc. Natl. Acad. Sci. U.S.A. 110, 12456–12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173).Hou X., Yoshioka N., Tsukano H., Sakai A., Miyata S., Watanabe Y., et al. (2017) Chondroitin sulfate is required for onset and offset of critical period plasticity in visual cortex. Sci. Rep. 7, 12646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174).Yoshioka N., Miyata S., Tamada A., Watanabe Y., Kawasaki A., Kitagawa H., et al. (2017) Abnormalities in perineuronal nets and behavior in mice lacking CSGalNAcT1, a key enzyme in chondroitin sulfate synthesis. Mol. Brain 10, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175).Miyata S., Nadanaka S., Igarashi M., Kitagawa H. (2018) Structural variation of chondroitin sulfate chains contributes to molecular heterogeneity of perineuronal nets. Front. Integr. Nuerosci. 12, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176).Sato Y., Watanabe N., Fukushima N., Mita S., Hirata T. (2011) Actin-independent behavior and membrane deformation exhibited by the four-transmembrane protein M6a. PLoS One 6, e26702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177).Ida-Yonemochi H., Morita W., Sugiura N., Kawakami R., Morioka Y., Takeuchi Y., et al. (2018) Craniofacial abnormality with skeletal dysplasia in mice lacking chondroitin sulfate N-acetylgalactosaminyltransferase-1. Sci. Rep. 8, 17134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178).Busch S.A., Silver J. (2007) The role of extracellular matrix in CNS regeneration. Curr. Opin. Neurobiol. 17, 120–127. [DOI] [PubMed] [Google Scholar]
- 179).Tran A.P., Warren P.M., Silver J. (2018) The biology of regeneration failure and success after spinal cord injury. Physiol. Rev. 98, 881–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180).Silver J., Miller J.H. (2004) Regeneration beyond the glial scar. Nat. Rev. Neurosci. 5, 146–156. [DOI] [PubMed] [Google Scholar]
- 181).Bradbury E.J., Moon L.D., Popat R.J., King V.R., Bennett G.S., Patel P.N., et al. (2002) Chondroitinase ABC promotes functional recovery after spinal cord injury. Nature 416, 636–640. [DOI] [PubMed] [Google Scholar]
- 182).Carulli D., Laabs T., Geller H.M., Fawcett J.W. (2005) Chondroitin sulfate proteoglycans in neural development and regeneration. Curr. Opin. Neurobiol. 15, 116–120. [DOI] [PubMed] [Google Scholar]
- 183).Shen Y., Tenney A.P., Busch S.A., Horn K.P., Cuascut F.X., Liu K., et al. (2009) PTPσ is a receptor for chondroitin sulfate proteoglycan, an inhibitor of neural regeneration. Science 326, 592–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184).Coles C.H., Shen Y., Tenney A.P., Siebold C., Sutton G.C., Lu W., et al. (2011) Proteoglycan-specific molecular switch for RPTPσ clustering and neuronal extension. Science 332, 484–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185).Lang B.T., Cregg J.M., DePaul M.A., Tran A.P., Xu K., Dyck S.M., et al. (2015) Modulation of the proteoglycan receptor PTPσ promotes recovery after spinal cord injury. Nature 518, 404–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186).Takeuchi K., Yoshioka N., Higa Onaga S., Watanabe Y., Miyata S., Wada Y., et al. (2013) Chondroitin sulphate N-acetylgalactosaminyl-transferase-1 inhibits recovery from neural injury. Nat. Commun. 4, 2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187).Basso D.M., Fisher L.C., Anderson A.J., Jakeman L.B., McTigue D.M., Popovich P.G. (2006) Basso Mouse Scale for locomotion detects differences in recovery after spinal cord injury in five common mouse strains. J. Neurotrauma 23, 635–659. [DOI] [PubMed] [Google Scholar]
- 188).Mikami T., Kitagawa H. (2013) Biosynthesis and function of chondroitin sulfate. Biochim. Biophys. Acta 1830, 4719–4733. [DOI] [PubMed] [Google Scholar]
- 189).Inatani M., Irie F., Plump A.S., Tessier-Lavigne M., Yamaguchi Y. (2003) Mammalian brain morphogenesis and midline axon guidance require heparan sulfate. Science 302, 1044–1046. [DOI] [PubMed] [Google Scholar]
- 190).Piper M., Anderson R., Dwivedy A., Weinl C., van Horck F., Leung K.M., et al. (2006) Signaling mechanisms underlying Slit2-induced collapse of Xenopus retinal growth cones. Neuron 49, 215–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191).Masu M. (2016) Proteoglycans and axon guidance: A new relationship between old partners. J. Neurochem. 139, 58–75. [DOI] [PubMed] [Google Scholar]
- 192).Fawcett J.W., Verhaagen J. (2018) Intrinsic determinants of axon regeneration. Dev. Neurobiol. 78, 890–897. [DOI] [PubMed] [Google Scholar]
- 193).Curcio M., Bradke F. (2018) Axon regeneration in the central nervous system: Facing the challenges from the inside. Annu. Rev. Cell Dev. Biol. 34, 495–521. [DOI] [PubMed] [Google Scholar]
- 194).Hunter A.M., Leuchter A.F., Power R.A., Muthén B., McGrath P.J., Lewis C.M., et al. (2013) A genome-wide association study of a sustained pattern of antidepressant response. J. Psychiatr. Res. 47, 1157–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195).Yukawa T., Iwakura Y., Takei N., Saito M., Watanabe Y., Toyooka K., et al. (2018) Pathological alterations of chondroitin sulfate moiety in postmortem hippocampus of patients with schizophrenia. Psychiatry Res. 270, 940–946. [DOI] [PubMed] [Google Scholar]
- 196).Gopal A.A., Ricoult S.G., Harris S.N., Juncker D., Kennedy T.E., Wiseman P.W. (2017) Spatially selective dissection of signal transduction in neurons grown on netrin-1 printed nanoarrays via segmented fluorescence fluctuation analysis. ACS Nano 11, 8131–8143. [DOI] [PubMed] [Google Scholar]
- 197).Baba K., Yoshida W., Toriyama M., Shimada T., Manning C.F., Saito M., et al. (2018) Gradient-reading and mechano-effector machinery for netrin-1-induced axon guidance. eLife 7, e34593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198).Cosker K.E., Segal R.A. (2014) Neuronal signaling through endocytosis. Cold Spring Harb. Perspect. Biol. 6, a020669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199).Winckler B., Mellman I. (2010) Trafficking guidance receptors. Cold Spring Harb. Perspect. Biol. 2, a001826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200).Tojima T., Kamiguchi H. (2015) Exocytic and endocytic membrane trafficking in axon development. Dev. Growth Differ. 57, 291–304. [DOI] [PubMed] [Google Scholar]