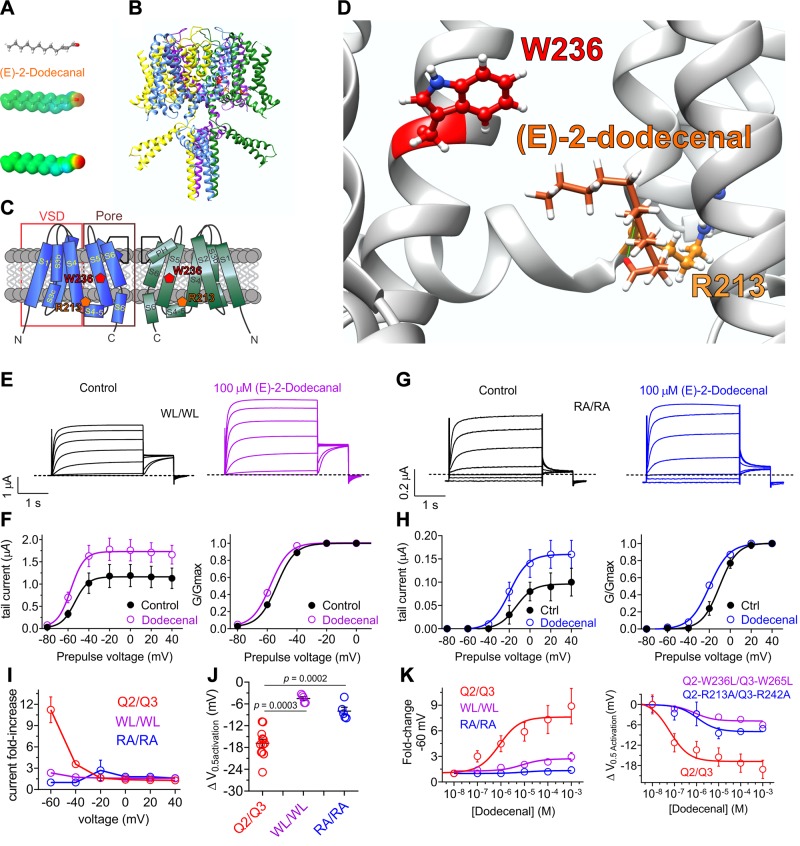
Figure 5.
KCNQ2/3 activation by (E)-2-dodecenal requires a conserved S5 tryptophan and S4–5 arginine. All error bars indicate sem. A) (E)-2-dodecenal chemical structure (upper and center) and electrostatic surface potentials (red, electron-dense; blue, electron-poor; green, neutral) (lower and center) calculated and plotted using Jmol. B) Chimeric KCNQ1/KCNQ2 structural model (orange, KCNQ2-R213; red, KCNQ2-W236). C) Topological representation of KCNQ5 showing 2 of the 4 subunits, without domain swapping for clarity. Pentagons, approximate position of KCNQ2-R213 (orange) and KCNQ2-W236 (red); D) View of the (E)-2-dodecenal binding site in KCNQ2 predicted by SwissDock. Green line, predicted H-bond. E) Mean TEVC current traces showing effects of (E)-2-dodecenal (100 µM) on KCNQ2-W236L/KCNQ3-W265L (WL/WL) channels expressed in Xenopus oocytes (n = 5–6). F) Mean tail current (left) and mean normalized tail currents (G/Gmax) (right) vs. prepulse voltage relationships for the traces as in E (n = 5–6). G) Mean TEVC current traces showing effects of (E)-2-dodecenal (100 µM) on KCNQ2-R213A/KCNQ3-R242 (RA/RA) channels expressed in Xenopus oocytes (n = 5–6). H) Mean tail current (left) and mean normalized tail currents (G/Gmax) (right) vs. prepulse voltage relationships for the traces as in G (n = 5–6). I) Current-fold increase vs. voltage in response to (E)-2-dodecenal (100 µM) of wild-type (Q2/Q3), WL/WL, and RA/RA KCNQ2/3 channels (n = 5–6). J) Scatter plot showing ΔV0.5 activation in response to (E)-2-dodecenal (100 µM) of wild-type (Q2/Q3), WL/WL and RA/RA KCNQ2/3 channels (n = 5–6). K) (E)-2-dodecenal dose response calculated from fold increase in current at −60 mV (left) and ΔV0.5 activation (right) for wild-type (Q2/Q3), WL/WL, and RA/RA KCNQ2/3 channels; n = 5–6.