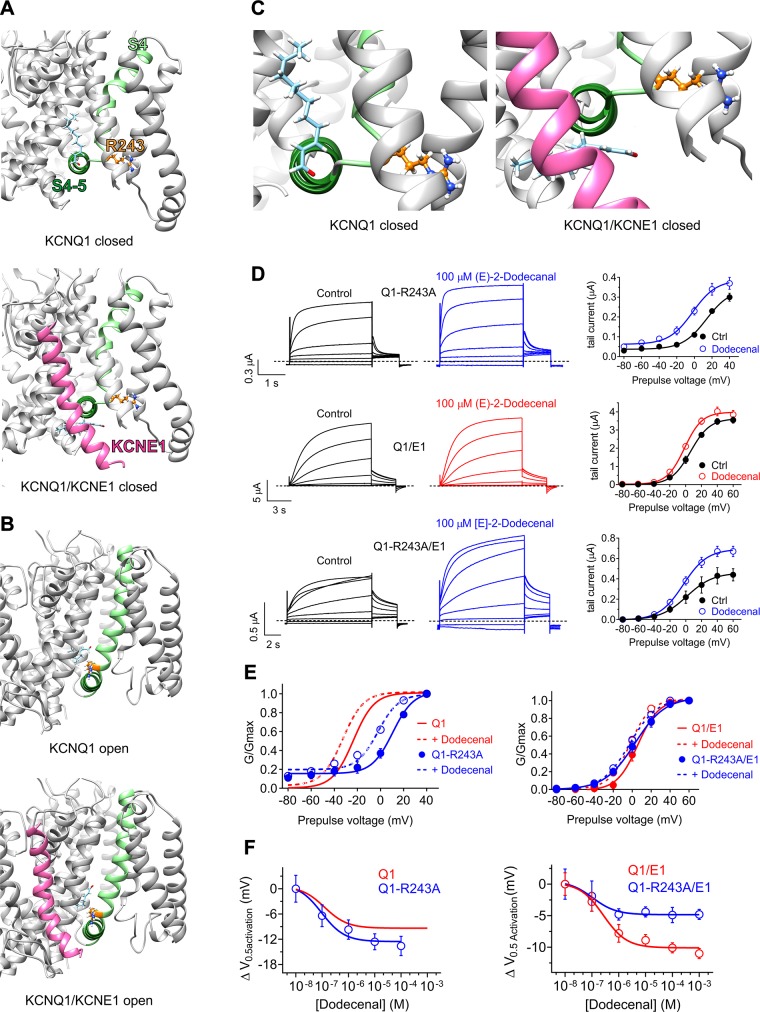
Figure 7.
KCNE1 impinges on the KCNQ1 (E)-2-dodecenal binding site. All error bars indicate sem. For structural models: Pink, KCNE1; pale green, KCNQ1-S4; forest green, KCNQ1-S4/5 linker; orange, KCNQ1-R243; pale blue, (E)-2-dodecenal. A) View of the (E)-2-dodecenal binding site predicted by SwissDock in the KCNQ1 (upper) and KCNQ1/KCNE1 (lower) closed-state models. B) View of the (E)-2-dodecenal binding site predicted by SwissDock in the KCNQ1 (upper) and KCNQ1/KCNE1 (lower) open-state models. C) Close-up view of the (E)-2-dodecenal binding site predicted by SwissDock (from A), highlighting the predicted KCNE1-induced shift in the (E)-2-dodecenal binding pose in the closed-state models. D) Mean TEVC current traces (left) and mean tail current vs. prepulse voltage relationships (right) showing effects of (E)-2-dodecenal (100 µM) on channels indicated (n = 5). E) Mean normalized tail current (G/Gmax) vs. prepulse voltage relationships showing effects of (E)-2-dodecenal (100 µM) (dashed lines) on the voltage dependence of activation of wild-type and R243A KCNQ1 (Q1) alone (left) or with KCNE1 (E1) (right); n = 5. Wild-type KCNQ1 data are from Fig 4B. F) (E)-2-dodecenal dose response calculated from ΔV0.5 activation for wild-type and R243A KCNQ1 (Q1) alone (left) or with KCNE1 (E1) (right); n = 5. Wild-type KCNQ1 data are from Fig. 4B.