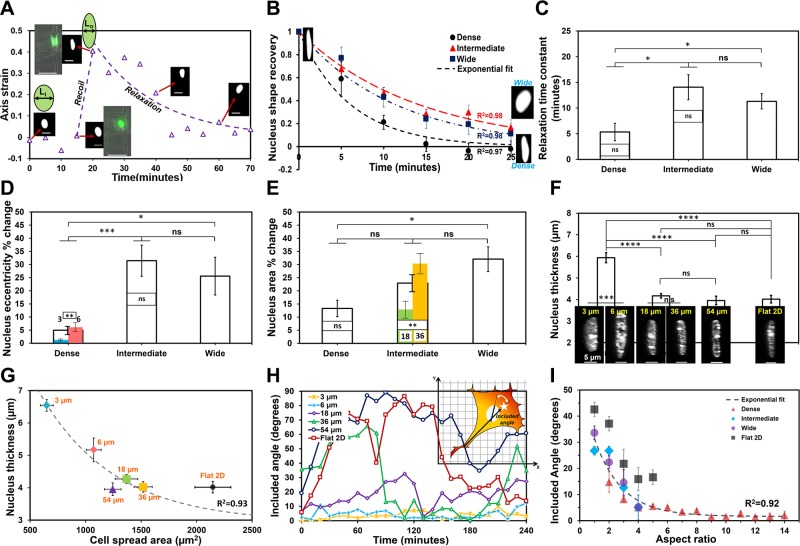
Figure 4.
Interfiber spacing modulates nucleus dynamics. A) Representative profile of the deformation of nucleus during retraction of a trailing edge demonstrating that rounded nucleus is strained to elongated shape, and, after recoil, the nucleus relaxes to its original shape. Axis strain is defined as ε = [Li – L(t)] ÷ Li, where Li is the minor axis length of the nucleus prerecoil and L(t) represents the minor axis length of the nucleus at time (t) (scale bars, 20 µm). Immediately postrecoil, minor axis length; L(t) = L0. B) Post-recoil normalized nucleus shape recovery data is defined as ε/ε0, where ε0 = (Li – L0) ÷ Li is the initial nucleus axis strain immediately postrecoil. Exponential fits to data show that cells on dense networks recover their original shape faster after recoil. Inset shows representative images of the nucleus postrecoil and after relaxation (rounded is for wide and elongated for dense networks). C) Relaxation time constants calculated on basis of an exponential decay of nucleus deformation showing stronger recoil in intermediate and wide networks. D, E) Percentage change in nucleus shape [eccentricity (D)] and size [projected area (E)] during trailing-edge recoil. F) Nucleus height measured by confocal microscopy quantitates cell nuclei to be thicker in dense regions. Inset shows representative images of nucleus (projected side view from confocal microscopy; scale bars, 5 µm). G) Nucleus thickness decreases with increase in cell spread area. H) Representative temporal plots of nucleus–cell body angular coupling (included angle) for the different interfiber spacing with inset illustration showing that the included angle is defined by the coordination between cell-body vector (trailing edge to centroid shown by white star) and nucleus major axis. I) Data show the influence of cell shape (aspect ratio) on the extent of nucleus–cell body coordination. For G and I, exponential fit to data corresponding to fiber networks with flat 2D data shown as reference. Sample size for B–E (n = 11, 23, and 10: dense, intermediate, and wide networks, respectively); for F and G (n = 25, 32, 14, and 13: dense, intermediate, wide, and flat 2D, respectively) and for I (n = 13, 15, 9, and 9: dense, intermediate, wide, and flat 2D, respectively). Included angle and aspect ratios are calculated every 10 min over the 4-h experimental time (25 measurements) and then averaged across cells of similar aspect ratios. Statistical differences between individual interfiber spacing categories are superimposed on the data for their respective groups; ns, not significant. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.