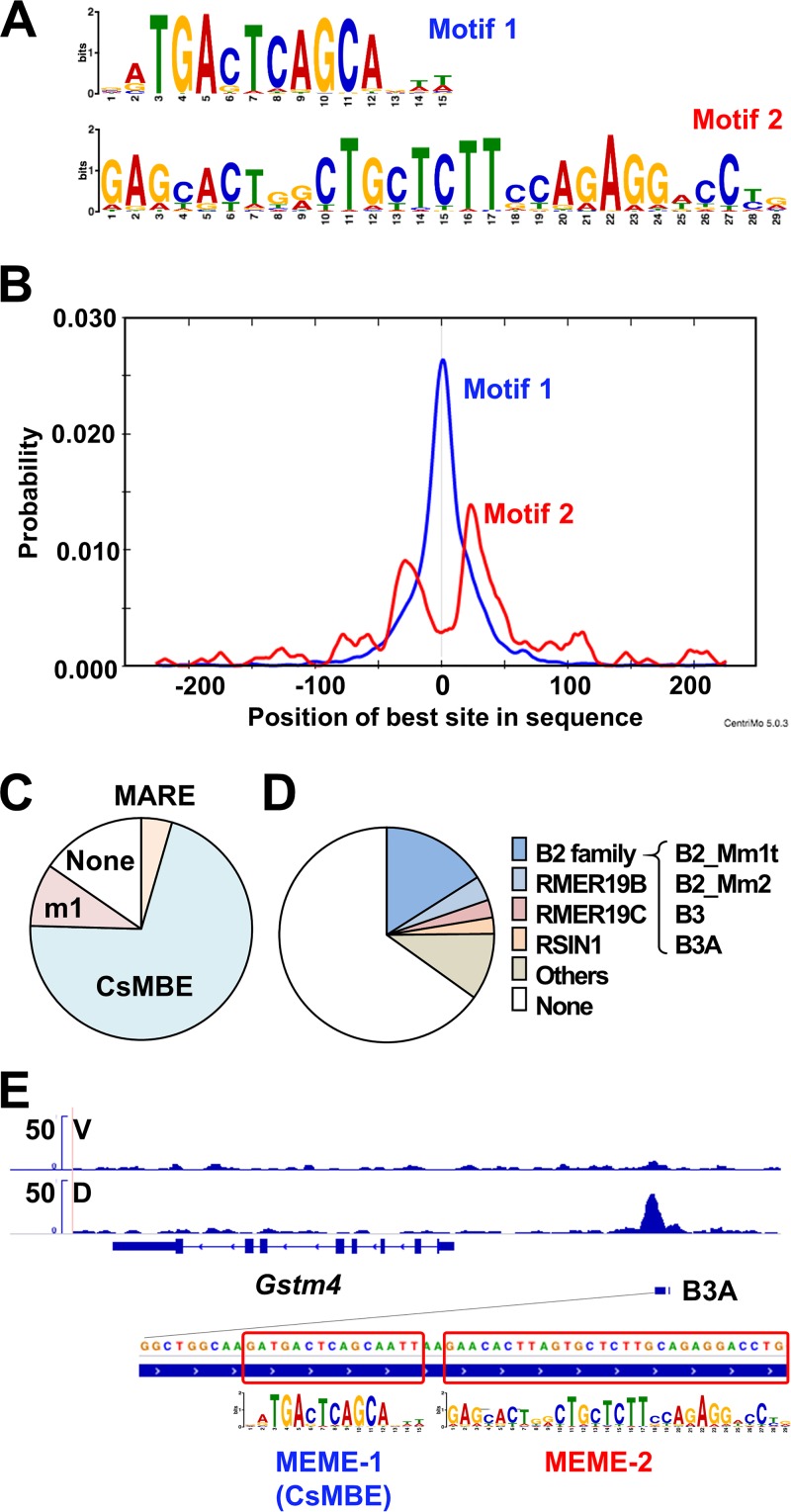
FIG 7.
De novo motif analysis of T-N2G binding sites. (A) Two motifs identified by the de novo motif discovery algorithm MEME-ChIP in the T-N2G binding sites are shown. (B) Average profiles of the enriched motifs in the T-N2G binding sites determined by MEME-ChIP. (C) Pie charts showing the proportions of MARE (GCTGASTCAGC), CsMBE (RTGASTCAGC), and m1 (one mismatch allowed in CsMBEs) in the T-N2G binding sites. (D) Pie charts showing the proportions of repeats that overlapped the T-N2G binding regions. Repeats deposited in the RepeatMasker data were used. (E) A B3A element, which belongs to the B2 SINE family and harbors a CsMBE, in the regulatory region of the Gstm4 gene. The locations of two enriched motifs are boxed.