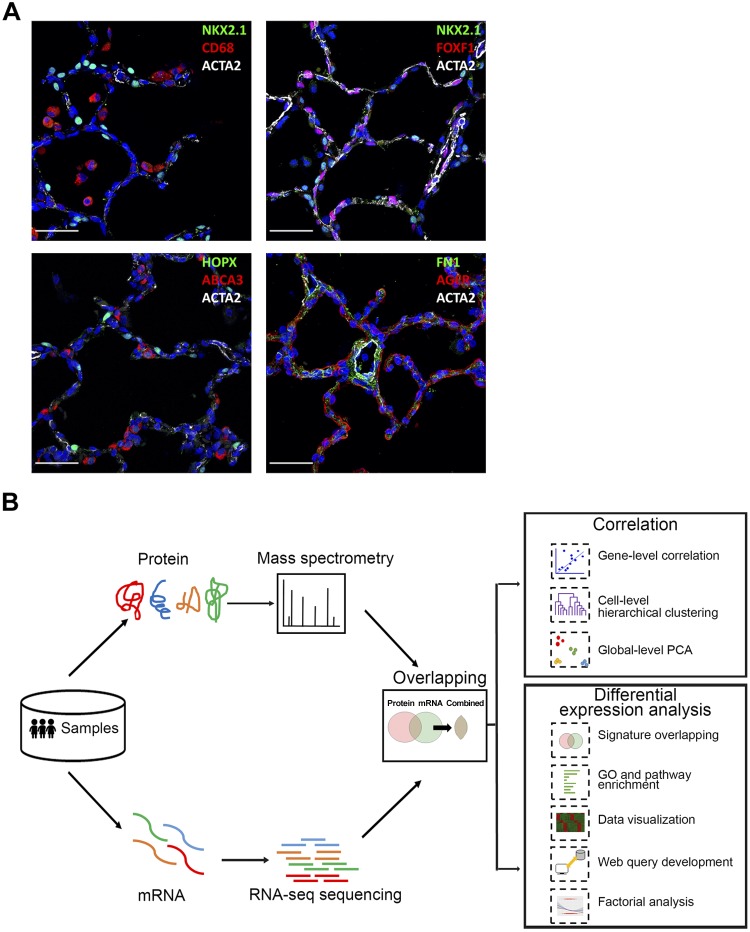
Fig. 1.
Anatomic location of human cell types and schematic of analytic workflow. A: confocal microscopy was utilized to define major lung cell types by immunofluorescence staining for NKX2-1, ABCA3 (AT2 cells), HOPX, AGER (AT1 cells), CD68 (immune cells), ACTA2 (smooth muscle/myofibroblast), FOXF1 (endothelial-mesenchymal cells), and FN1 (fibroblast) in donor lungs (D011 and D08) from infants of 20 mo of age. Scale bars, 100 μm. B: workflow of integrative transcriptomic and proteomic profiling analysis. Human lung samples underwent same sorting procedures to isolate endothelial, epithelial, immune, and mesenchymal cells for RNA-seq and proteomic profiling. 3,320 mRNA-protein pairs were obtained for downstream analyses (n = 3/cell type). Sample-level and gene-level correlations were calculated to assess the overall correlation of transcriptomic and proteomic data; differential RNA/protein expression analyses were performed using one-way ANOVA with REML (restricted maximum likelihood) model and compared to identify coherent vs. noncoherent signatures for major cell types; functional enrichment analyses were performed to identify unique and shared biological processes and pathways; and chi-square test and logistic regression analysis were conducted to determine potential factors influencing mRNA-protein expression coherency. Data visualization and web query tools were developed to facilitate data sharing and reutilization. PCA, principle component analysis; GP, gene ontology.