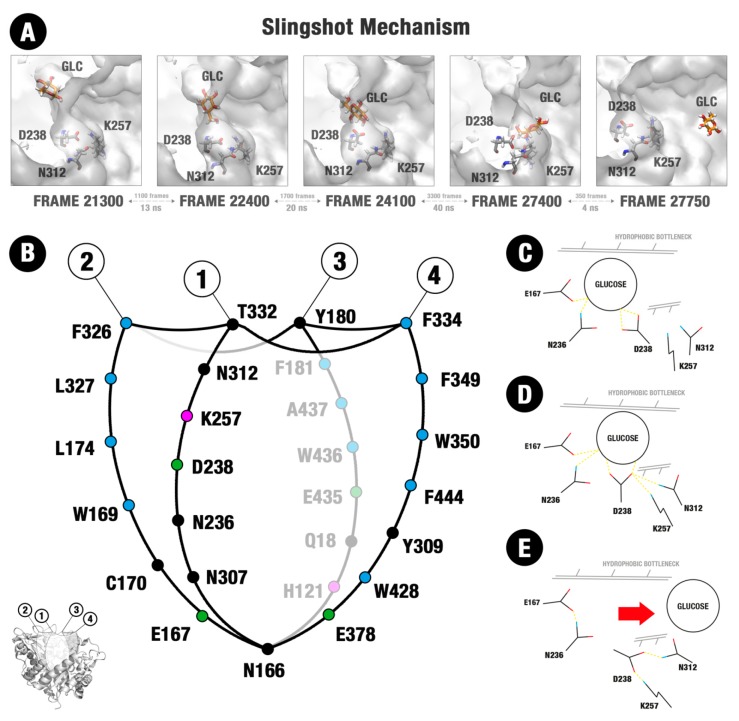
Figure 6.
Structure of the substrate channel of HiGB and the mechanism of glucose release. (A) Five snapshots of the MD simulation that illustrate the slingshot mechanism and residues involved. (B) Structure of the substrate channel organized into four sectors (1–4). A set of hydrophobic residues is located in the entrance of this channel, mainly in the sectors 2, 3, and 4. Circles represent apolar (blue), polar neutral (black), polar positively charged (magenta), and polar negatively charged (green) amino acids. (C–E) Amino acid residues involved in the slingshot mechanism: E167, N236, D238, K257, and N312.