Figure 1.
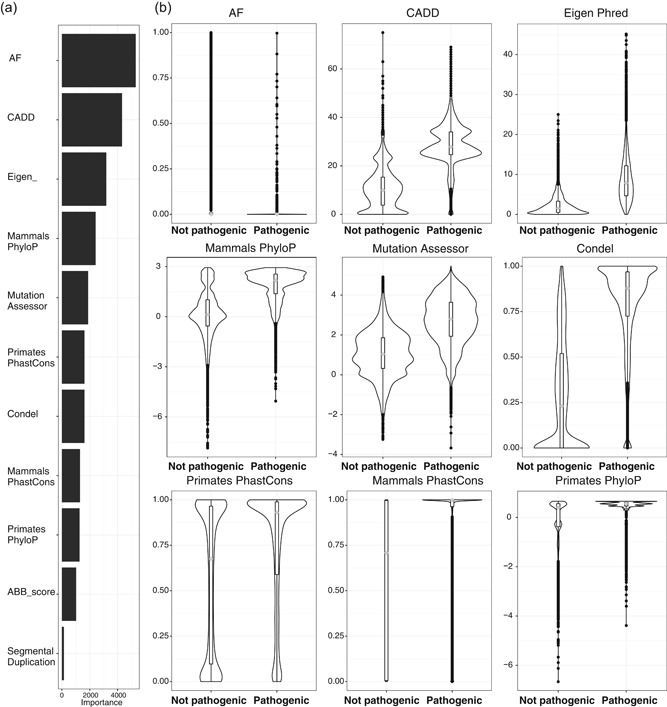
eDiVA‐Score random forest model. (a) Estimated importance of features used in the model (extracted with varImp command). (b) Distribution of values for the top‐9 features used in the model, comparing ClinVar pathogenic against ClinVar nonpathogenic variants. AF: allele frequency; eDiVA: exome Disease Variant Analysis
