Figure 3.
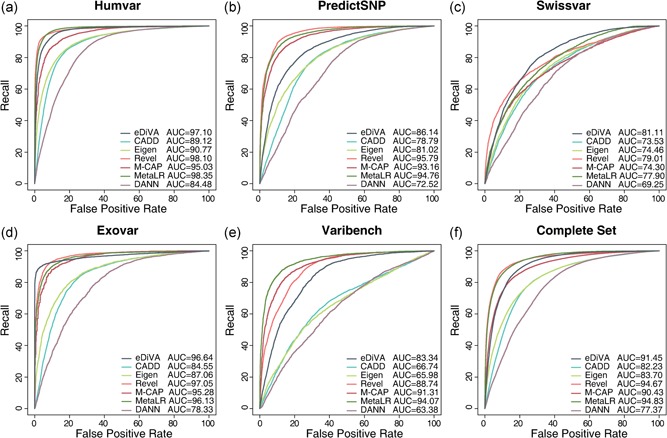
Receiver operating characteristic curves comparing pathogenicity classifiers on five independent data sets (and the combined set) composed of pathogenic and neutral variants. Revel, M‐CAP, and eDiVA show a similarly strong performance, with the exception of the PredictSNP and Varibench sets, on which Revel and M‐CAP outperform eDiVA‐Score. eDiVA: exome Disease Variant Analysis; M‐CAP: Mendelian clinically applicable pathogenicity; SNP: single‐nucleotide polymorphism
