Figure 4.
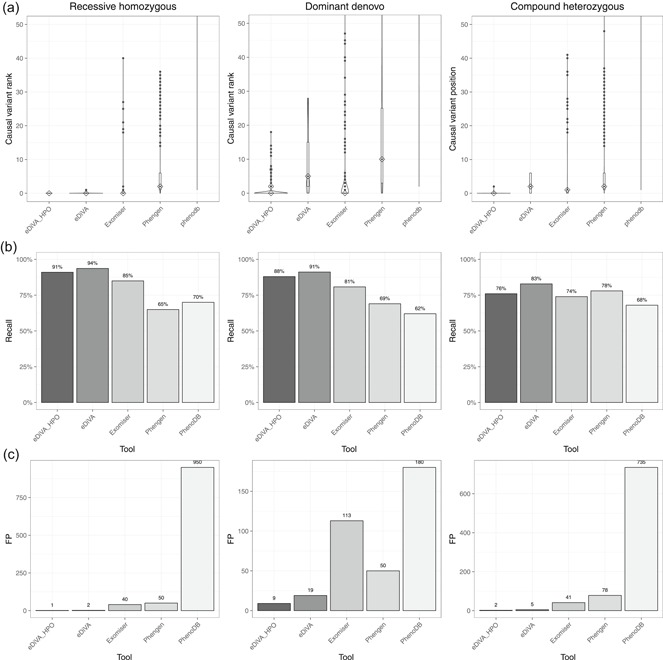
Benchmark of the causal variant prioritization tools eDiVA, Exomiser, Phen–Gen, and PhenoDB. (a) Violin plots showing the rank of disease‐causing variants within the reported candidate lists for the three tested inheritance types: “recessive homozygous”, “compound heterozygous”, and “dominant de novo”; (b) Recall values for 6,811 semisynthetic trio cases, representing the fraction of identified causal variants (i.e., “solved cases”). (c) Average number of false positives reported per case as a proxy for precision. eDiVA has been tested in two configurations, with HPO‐based gene prioritization (eDiVA_HPO) and with the default configuration not using HPO terms (eDiVA). Adding HPO filtering reduces false positives at the cost of a slightly reduced Recall. HPO: Human Phenotype Ontology
