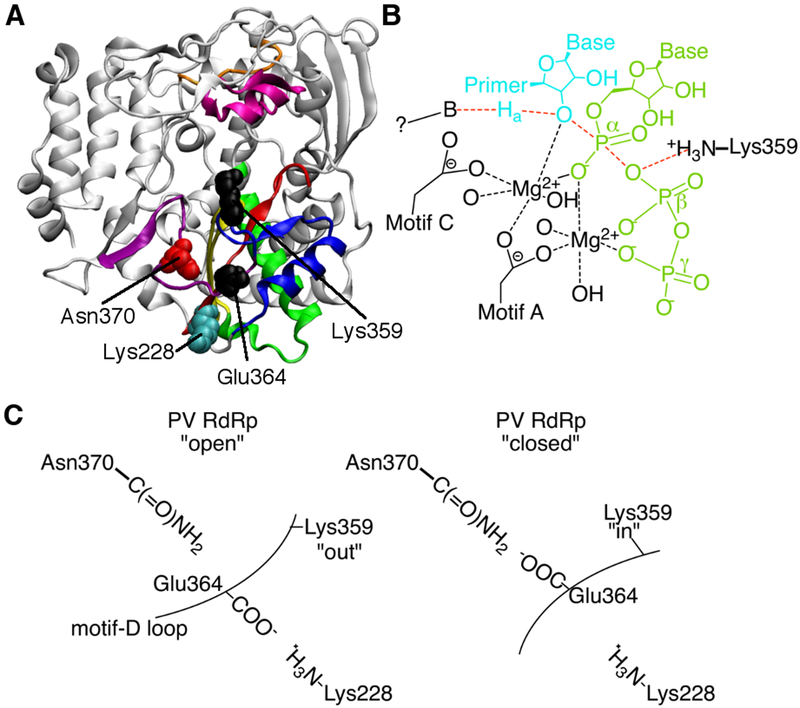
Figure 1.
Motif-D loop interactions in the poliovirus RNA-dependent RNA polymerase. (A) The X-ray crystal structure of PV RdRp53 (PDB 1RA6) showing conserved structural motifs colored as followed: A, red; B, green; C, yellow; D, blue; E, purple; F, pink. Key residues on and making interactions with motif D are indicated. (B) Chemical mechanism of PV RdRp indicating the general acid role of Lys359 to protonate the leaving pyrophosphate group. The “?” represents a proposed general base that has yet to be identified. (C) Simplified model showing the interactions made by loop residue Glu364 in the “open” and “closed” states of PV RdRp. In the “open” state, Glu364 makes an interaction with Lys228 and Lys359 is positioned away from the active site, unable to serve as the general acid. In the “closed” state, Glu364 makes an interaction with Asn370 and Lys359 is repositioned such that it can act as the general acid.