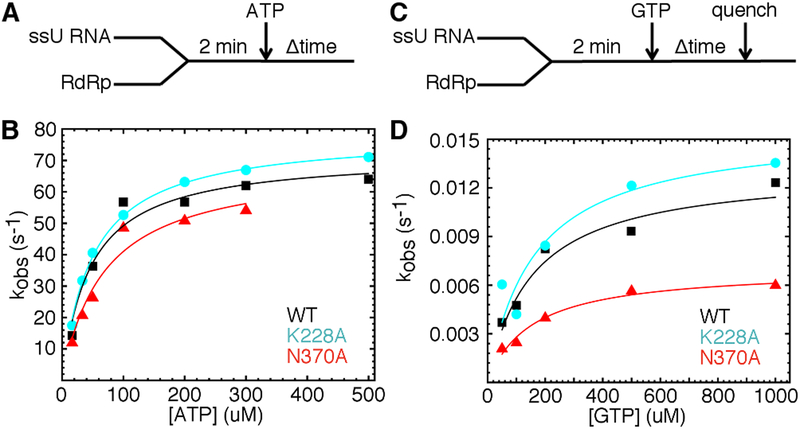
Figure 3.
The K228A and N370A substitutions lead to lower and higher fidelity polymerases respectively. (A) Experimental design for single nucleotide incorporation assays using ATP and 2’-dATP. RdRp (1 μM) was pre-incubated with ssU RNA (1 μM) before being quickly mixed with equal volume of ATP or 2’-dATP with different concentrations. These reactions were monitored by fluorescence changes over time using a stopped-flow apparatus. (B) Kinetic data for correct AMP incorporation (templated against U) for WT (black), K228A (cyan) and N370A (red) PV RdRp. The lines represent data fit to a hyperbola function to give an apparent dissociation constant (Kd,app) and a maximal rate constant for nucleotide incorporation (kpol). Kinetic parameters were determined at least three times; data shown represents one set of experiments. (C) Experimental design for single nucleotide incorporation assays using GTP. RdRp (1 μM) was pre-incubated with ssU RNA (1 μM) at room temperature for 3 min and then at the assay temperature of 30 °C for 2 min, before being quickly mixed with equal volume of GTP at different concentrations. In this case, RNA was 32P labeled on the 5′-end. (D) Kinetic data for incorrect GMP incorporation (templated against U) for WT (black), K228A (cyan) and N370A (red) PV RdRp. The lines represent data fit to a hyperbola function to give an apparent dissociation constant (Kd,app) and a maximal rate constant for nucleotide incorporation (kpol). Kinetic parameters were determined at least three times; data shown represents one set of experiments.