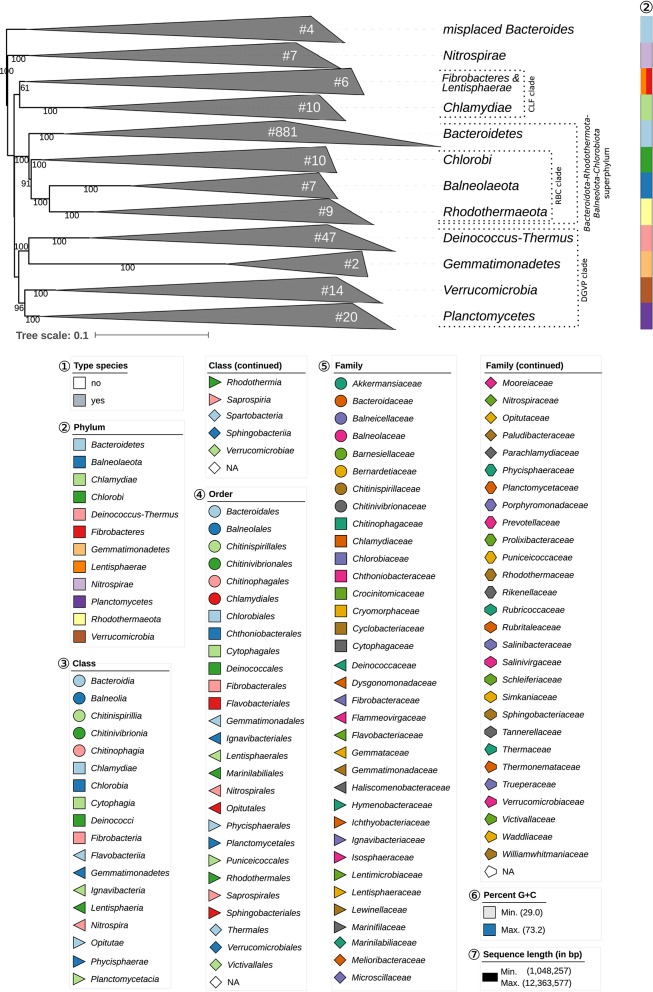
Figure 1.
Overview of the phylogenomic tree inferred with FastME from GBDP distances calculated from whole proteomes. The numbers above branches are GBDP pseudo-bootstrap support values from 100 replications. Collapsed clades are displayed as triangles whose side lengths are proportional to the branch-length distances to least and most distant leave, respectively. The total number (#) of leaves per collapsed clade is shown within the triangles. The legend indicates the symbols and colors used in all subsequent figures, which show details of all clades of interest. These clades are composed of the following phyla: CLF, Chlamydiae-Lentisphaerae-Fibrobacteres clade; DGPV, Deinococcus-Thermus-Gemmatimonadetes-Planctomycetes-Verrucomicrobia clade; RBC, Rhodothermaeota-Balneolaeota-Chlorobi clade. These clades are weakly supported and annotated for display purposes only; they are not suggested as reliable groupings. Figures 2–8 show specific sections of the same tree in greater detail; while the underlying topology is exactly the same, the ordering of the clades may slightly differ.