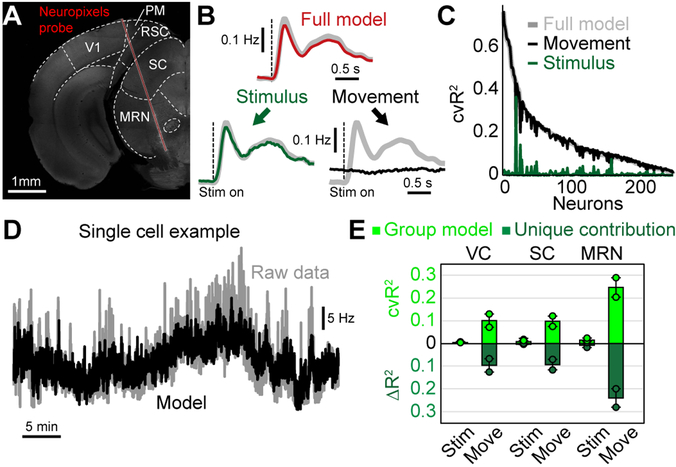
Figure 8. Uninstructed movements predict single-neuron activity in cortical and subcortical areas.
(A) Coronal slice, −3.8 mm from bregma, showing the location of the Neuropixels probe in an example recording. We recorded activity in visual cortex (VC), superior colliculus (SC) and the midbrain reticular nucleus (MRN). (B) Population PETH averaged over all neurons and visual stimuli, n = 232 neurons. Gray traces show recorded data. Top red trace shows model reconstruction. Bottom: PETH partitioning of modeled data into stimulus (left, green trace) and movement (right, black trace) components. (C) Explained variance for all recorded neurons using either the full model (gray) or a movement- (black) or stimulus-only model (green). (D) Activity of a single example neuron over the entire session (~40 minutes). Gray trace is recorded activity, black is the cross-validated model reconstruction. (E) Explained variance for stimulus or movement variables in different brain areas. Shown is either all explained variance (light green) or unique model contribution (dark green). Bars represent the mean from 2 mice for VC, SC and MRN, respectively. Dots indicate the mean for each animal (on average 38.7 neurons per mouse and area). Error bars show the SEM.