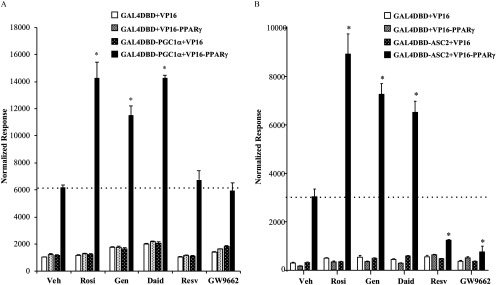
Figure 7.
The interaction of with the transcriptional coactivators and ASC-2 in the presence of agonist rosiglitazone, phytoestrogens, or the antagonist GW9662. HeLa cells were transiently transfected with and (A) or (B) Gal4DBD-ASC-2 expression plasmid or empty controls (VP16, Gal4DBD) together with a 5xGAL4 luciferase reporter and a luciferase normalization control. Following transfection, cells were treated with vehicle (Veh) or of Rosiglitazone (Rosi), Genistein (Gen), Daidzein (Daid), Resveratrol (Resv), or antagonist GW9662. After 36 h, cells were harvested and Dual-Luciferase assays were performed. Each value was normalized to the internal luciferase control. Each data point is the average of four independent experiments; individual experiments included three technical replicates used for each treatment. Bars represent . The dotted line on each graph indicates the basal level of coactivator interaction with for comparison with ligand-mediated interactions. * for comparison between Veh and each treatment.