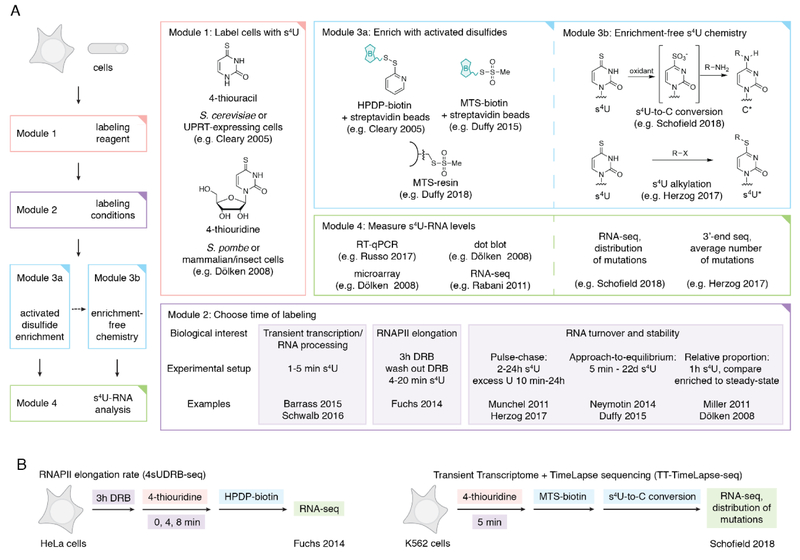
Figure 1:
(A) Overview of s4U metabolic labeling experiments. Different options are indicated for each module, along with references for representative examples. In the case where two references are listed, the first applies to yeast and the second to mammalian cells, as the time of s4U labeling can vary widely between organisms. (B) Representative examples of two s4U metabolic labeling experiments that leverage different module options: 4sUDRB-seq to measure RNAPII elongation and TT-TimeLapse-seq to detect unstable RNAs.