Figure 5.
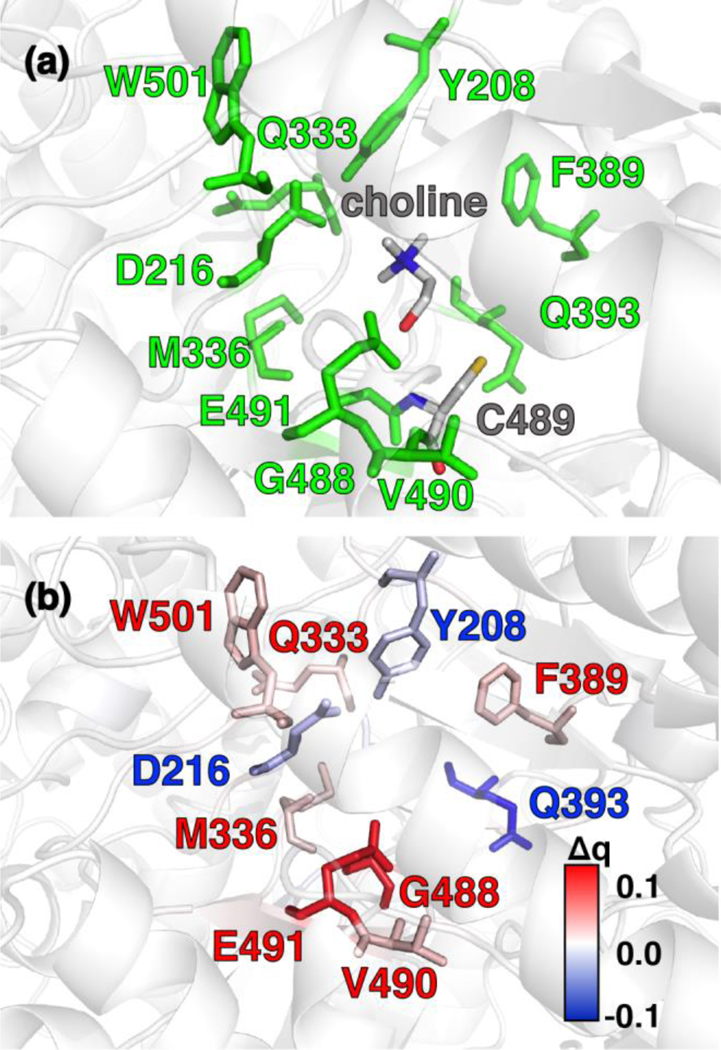
(a) CutC active site structure with choline substrate and cysteinyl radical (C489) shown in gray. Protein residues that were identified by charge shift analysis of QM regions are shown in green. (b) Difference of residue VDD charge upon substrate removal of the choline and residue mutation of C489 to alanine. Residues color-coded in blue and red correspond to loss and gain of partial charge upon substrate removal and residue mutation, respectively (as shown in inset color bar). All residues with Δq ≥ |0.015| e are shown as sticks. Residues of interest are labeled by their single letter residue code and number.
