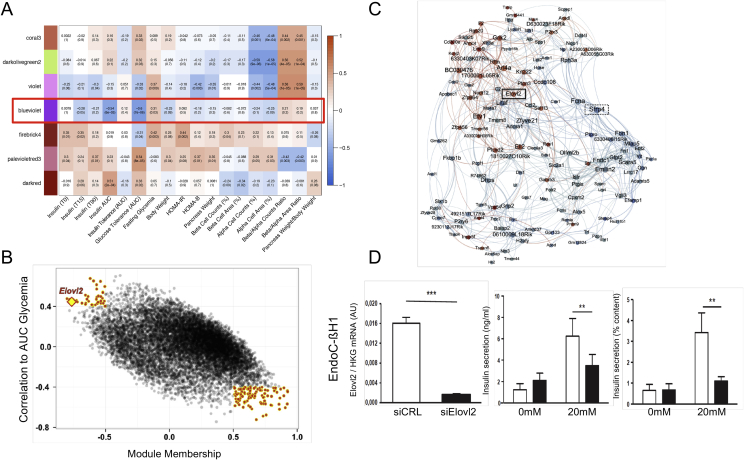
Figure 4.
A gene co-expression module correlated to insulin secretion and oral glucose intolerance. (A) Heat map showing correlations between module eigengenes and mouse phenotypic traits: darker colors indicate higher Spearman correlation. The red box indicates the correlations corresponding to the blue-violet module. (B) Scatter plot of AUC glycemia correlation against module membership (correlation to module) for all genes of the blue-violet module. Genes with the strongest correlations to both the module and to AUC glycemia are highlighted by red points. Elovl2 is indicated by a yellow diamond. (C) Network generated from the selected module genes. Node size is proportional to degree and node color indicates correlation to AUC glycemia (blue: negative correlation; red: positive correlation). Edges (connections) between nodes indicate correlation between genes (blue: negative; red: positive). Elovl2 and Sfrp4 are indicated in the network. (D) Effects of Elovl2 loss of function on glucose-stimulated insulin secretion in the human EndoC-βH1 cell line. Left: Elovl2 mRNA silencing; middle: insulin secretion expressing in ng/ml; right: insulin secretion as % of content. Values are mean (±SE) of three independent experiments. ***p < 0.001; **p < 0.01; *p < 0.05. Panels reproduced from [13].