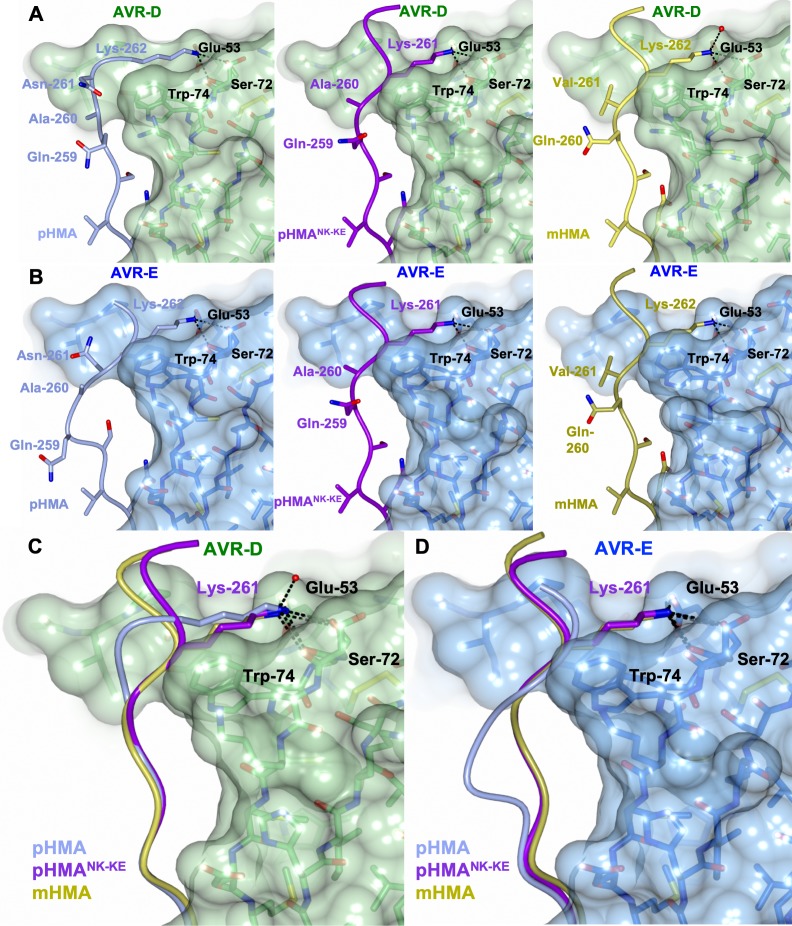
Figure 3. The PikpNK-KE-HMA mutant adopts a Pikm-like conformation at the effector-binding interface.
Schematic view of the different conformations adopted by Pikp-HMA, Pikp-HMANK-KE and Pikm-HMA at interface 3 when in complex with AVR-PikD or AVR-PikE. In each panel, the effector is shown as sticks with the molecular surface also shown and colored as labeled. Pik-HMA residues are colored as labeled and shown in the Cα-worm with side-chain representation. (A) Schematic of Pikp-HMA (left), Pikp-HMANK-KE (middle) and Pikm-HMA (right) bound to AVR-PikD. Important residues in the HMA–effector interaction are labeled as shown. (B) Schematic of HMA residues as for panel (A), but bound to AVR-PikE. (C) Superposition showing Pikp-HMA, Pikp-HMANK-KE and Pikm-HMA chains (colored in blue, purple and yellow, respectively) bound to AVR-PikD. For clarity, only the Lys-261/262 side chain is shown. (D) Superposition as described before, but bound to AVR-PikE.