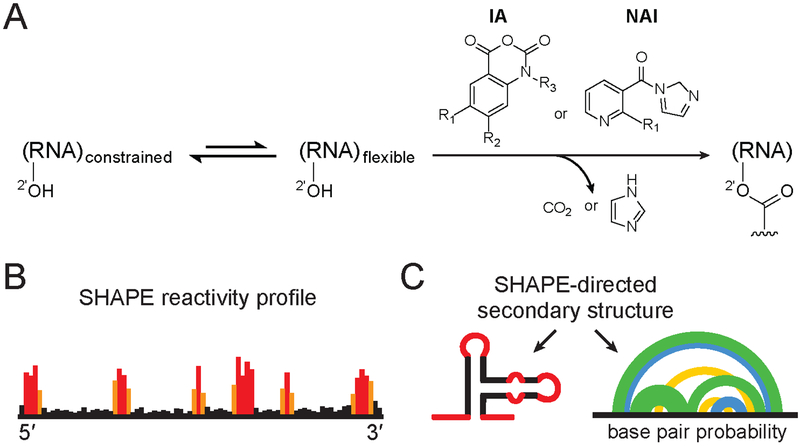
Figure 1.
SHAPE chemistry mechanism, reagents used to probe viral RNA structure, and SHAPE-directed structure modeling. (A) SHAPE reagents react with the 2′-hydroxyl group of conformationally flexible RNA nucleotides, yielding a covalent 2’-O-adduct. Widely used reagents are based on the isatoic anhydride (IA) or nicotinic acid imidazolide (NAI) scaffolds (47, 54, 115). (B) Representative SHAPE reactivity profile, with black, orange and red bars indicating low, medium and high nucleotide reactivities. (C) Use of SHAPE data to model a single minimum free energy structure (left) or characterize an ensemble consistent with SHAPE data (right). Arcs (right) connect base paired nucleotides with pairing probabilities illustrated (from highest to lowest) in green, blue and yellow. Data in panels B and C correspond to the same representative structural element; adapted from Siegfried et al. (37).