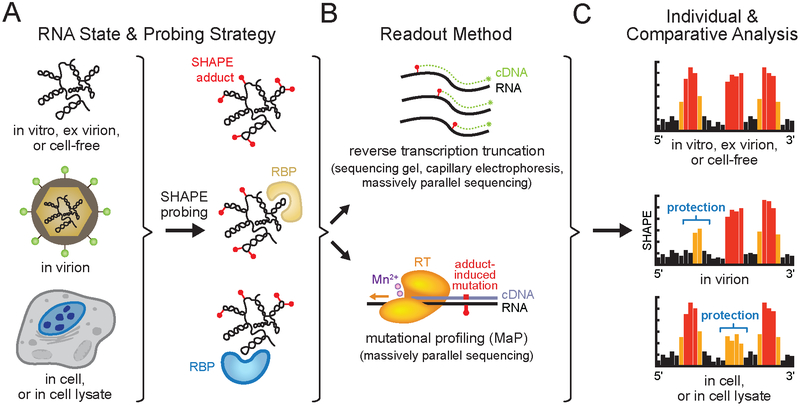
Figure 2.
SHAPE probing strategies. (A) Chemical probing of a viral RNA genome in an informative biological state (see Box 1 for definitions). Representative SHAPE adducts (red spheres) and RNA binding proteins (RBP) are shown. (B) Readout of per-nucleotide SHAPE reactivities by adduct-induced reverse transcription primer extension truncation or mutational profiling (MaP) and quantified by sequencing gel, capillary electrophoresis or massively parallel sequencing. (C) SHAPE reactivity profiles from distinct biological states can be compared to identify sites of SHAPE reactivity protections and enhancements, revealing state-specific RNA conformations, or protein or small molecule binding. Black, orange and red bars indicate low, medium and high nucleotide reactivities, respectively. Individual SHAPE reactivity profiles can also be incorporated into modeling algorithms to yield RNA secondary structure models (see Figure 1). Elements of figure adapted from Smola et al. (38, 116).