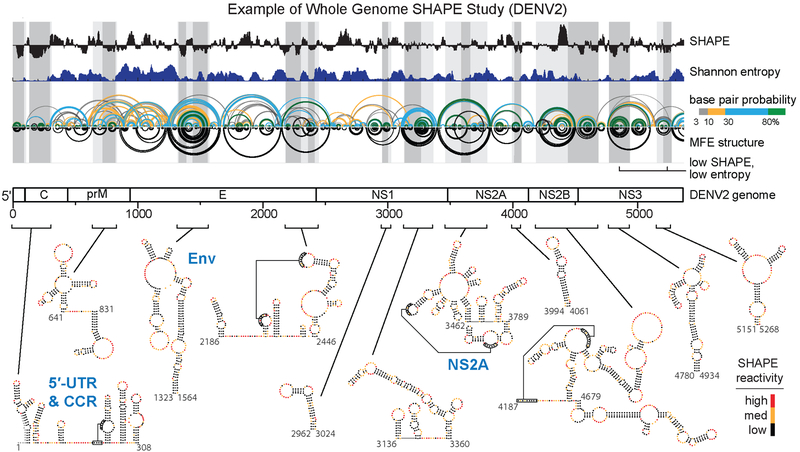
Figure 3.
Whole viral genome studies and well-determined structures. Genome wide SHAPE structure probing from a representative study of the DENV2 RNA genome. The 5’ half of the ~11,000 nucleotide long RNA genome is shown. Median ex virion 1M7 SHAPE reactivities (black) and Shannon entropies (dark blue) are plotted over centered 55-nt windows. Regions with both low SHAPE and low Shannon entropy, termed low SHAPE, Shannon entropy regions, are highlighted by dark-gray shading, with light-gray shading extended to encompass entire structures. Base pairing arcs are colored by probability (see scale), with green arcs indicating the most probable and well-determined base pairs. The minimum free-energy (MFE) secondary structure (inverted black arcs) was obtained using SHAPE reactivities as restraints (37, 38, 117). Secondary structures of the twelve boxed low SHAPE, low Shannon entropy elements are shown at bottom of image and are colored by SHAPE reactivity. Previously studied RNA structural elements in the 5′-UTR and capsid-coding region (CCR) and elements correctly modeled de novo by SHAPE-directed modeling are labeled in blue. Novel structural elements in the Env- and NS2A- coding regions identified by SHAPE structure probing and shown to be critical for viral fitness are also labeled in blue. Adapted from Dethoff et al. (58).