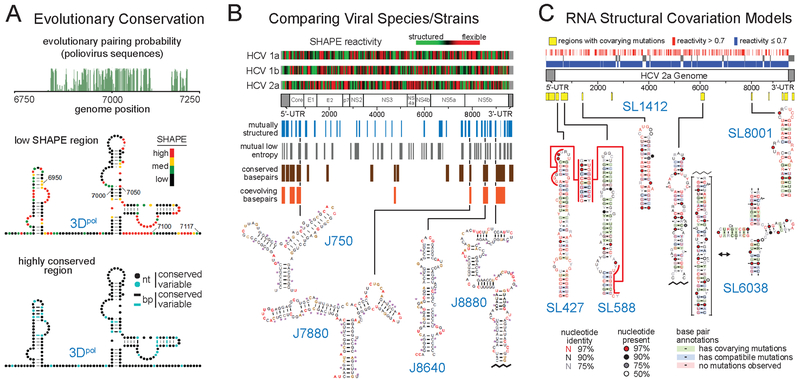
Figure 4.
Representative methods for melding sequence covariation information with SHAPE-directed structure models to identify functional RNA elements. (A) Coupling evolutionary pairing probability analysis of viral sequence alignments (green) with SHAPE structure probing to identify the 3Dpol functional element in the poliovirus genome. RNA nucleotides are labeled with SHAPE reactivities or nucleotide (nt) and base pair (bp) conservation. Adapted from Burrill et al. (78) with permission from American Society for Microbiology. (B) Comparison of sequences, SHAPE reactivities, and structure models across three HCV genotype subtypes to identify conserved functional structural elements. RNA structures are labeled by position in the JFH1 strain and residues are colored by SHAPE reactivity. Adapted from Mauger et al. (65). (C) RNA structural covariation models, built from a SHAPE-directed HCV secondary structure model and >1000 divergent HCV sequences. Functional motifs are colored by structural consensus and covariation. Conserved elements are located at different positions in distinct HCV genome sequences. Adapted from Pirakitikulr et al. (69) with permission from Elsevier.