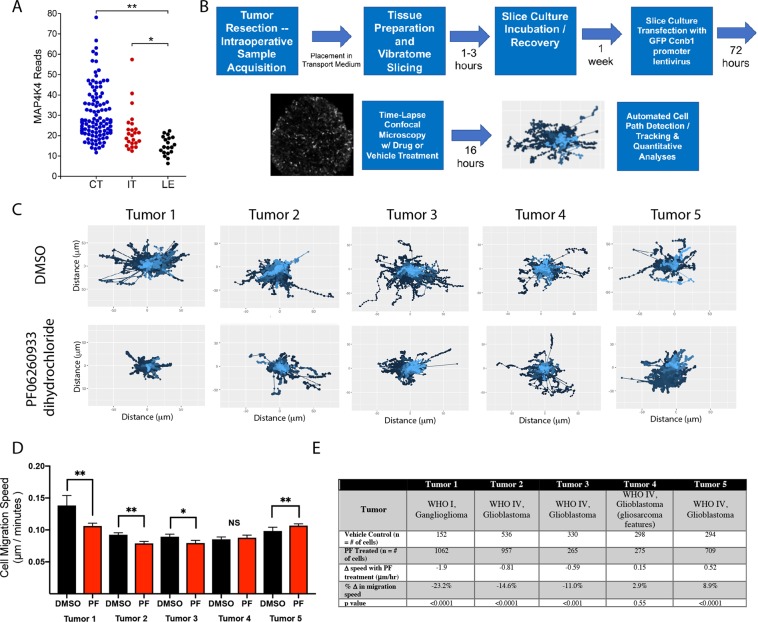
Figure 5.
MAP4K inhibition decreases tumor cell migration in a subset of primary human glioma derived organotypic slice cultures. (A) MAP4K4 expression in tumor samples from the IVY Glioblastoma Atlas http://glioblastoma.alleninstitute.org/16. Each point corresponds to an individual laser-microdissected sample. Samples are organized by tumor sub-region, including cellular tumor (CT), infiltrating tumor (IT), and leading edge (LE), (*p < 0.002; **p < 6 × 10−9 rank sum test). No multiple-comparison corrections were applied. (B) Experimental workflow schematic for organotypic slice cultures outlining tissue acquisition, slicing, transfection, time-lapse imaging, and cell migration analyses. (C) Displacement maps from representative slice culture imaging regions from vehicle control (DMSO) top and PF06260933 dihydrochloride treated (bottom) for all 5 patient tumor samples highlight variability in baseline cell dispersion and efficacy of MAP4K4 inhibition. (D) Individual cell migration paths were detected by ADAPT and cell migration speed calculated using a distance over time method. Mean cell speeds from vehicle treated slice regions (black) and PF treated (red) for each tumor. Error bars represent 95% confidence interval (**p < 0.0001; *p < 0.001 Mann-Whitney). Individual cell migration speeds were aggregated from multiple tumor slices to capture population level effects of MAP4K4 inhibition and groups were compared using the Mann-Whitney U Test given the non-normality of cell migration speed distributions. Significance was defined as p < 0.05. (E) Description of tumor pathology, number of cells tracked in the vehicle control and PF06260933 dihydrochloride treated regions and change in mean migration speed of the population.