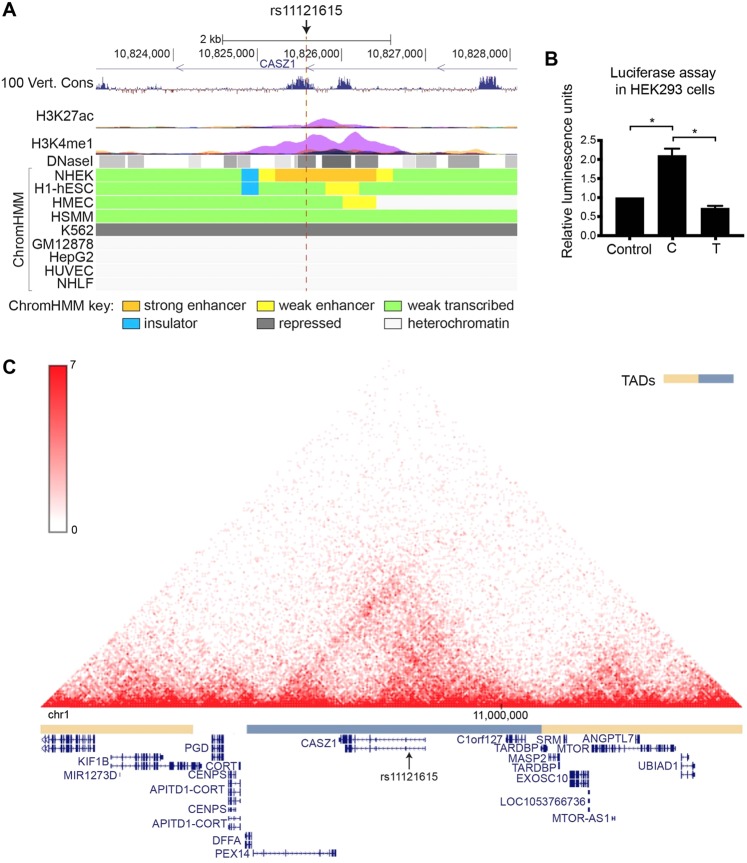
Figure 4.
rs11121615 affects enhancer activity within a topologically associated domain (TAD) containing CASZ1. (A) UCSC snapshot showing the following tracks: UCSC genes, 100 vertebrates basewise conservation by PhyloP, ENCODE layered ChIP-Seq data for H3K27ac and H3K4me1 from seven cell lines (NHEK cells in purple), ENCODE DNase I hypersensitivity data, and 15-state chromatin state Segmentation by HMM (ChromHMM) for nine cell lines. hg19 chromosome 1 coordinates are shown. (B) Allele-specific enhancer activity for rs11121615 was assessed using a luciferase reporter assay in HEK293 cells. The rs11121615 T variant (major allele) did not harbour enhancer activity whereas the C variant (minor allele) did. The average of five biological replicates+/− the standard error of the mean is shown. Statistical significance was determined by a one-way ANOVA followed by a Tukey’s multiple comparisons test (*P < 0.0001). (C) Hi-C contact map in NHEK cells (normal human epidermal keratinocyte) at chr1:10140000-11450000 (hg19), obtained from the 3D genome browser (http://www.3dgenome.org). TADs, calculated using the directional bias method13, are indicated by alternating orange and blue bars. NCBI reference genes are annotated below.