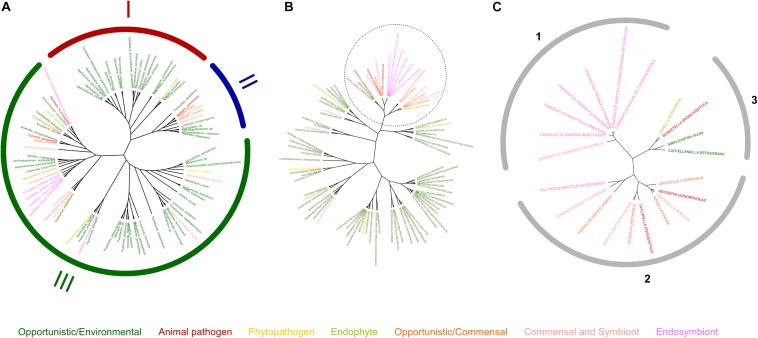
FIGURE 5.
Hierarchical clustering of β proteobacteria based on their copper-related proteomes. (A) Phylogenetic tree based on the 16S RNA sequences. For readability, the branches of the tree do not represent true phylogenetic distances, and only one species of Burkholderia (B. cepacia), Bordetella (B. bronchiseptica), and Neisseria (N. gonorrhoeae) were used in these analyses. The tree thus comprises 86 species that form three large phylogenetic groups, namely Rhodocyclales and Nitrosomonadales (I), Nesseriales (II), and Burkholderiales (III). (B) Tree representation of the hierarchical clustering of the 86 bacterial species based on their sets of copper-homeostasis proteins (composition and numbers). The color code is as follows: dark green, environmental and opportunistic/environmental bacteria; pale green, endophytes; yellow, phytopathogens; salmon pink, commensal bacteria and symbionts; pink, endosymbionts; orange, opportunistic/commensal bacteria; red, animal pathogens. (C) The upper branch of the tree shown in (B) (21 species) was further analyzed by hierarchical clustering based on the complete sets of copper-related proteins in the bacteria present in that branch. The results are represented as a tree. The three groups that result from this analysis are (1) bacteria fully dependent on host cells (endosymbionts); (2) extracellular bacteria that depend on hosts; and (3), environmental bacteria. See text for details.